沉默减数分裂内切酶1(EME1)抑制肝癌细胞增殖的机制
DOI: 10.12449/JCH240518
Silencing essential meiotic endonuclease 1 inhibits the proliferation of liver cancer cells: A study of related mechanisms
-
摘要:
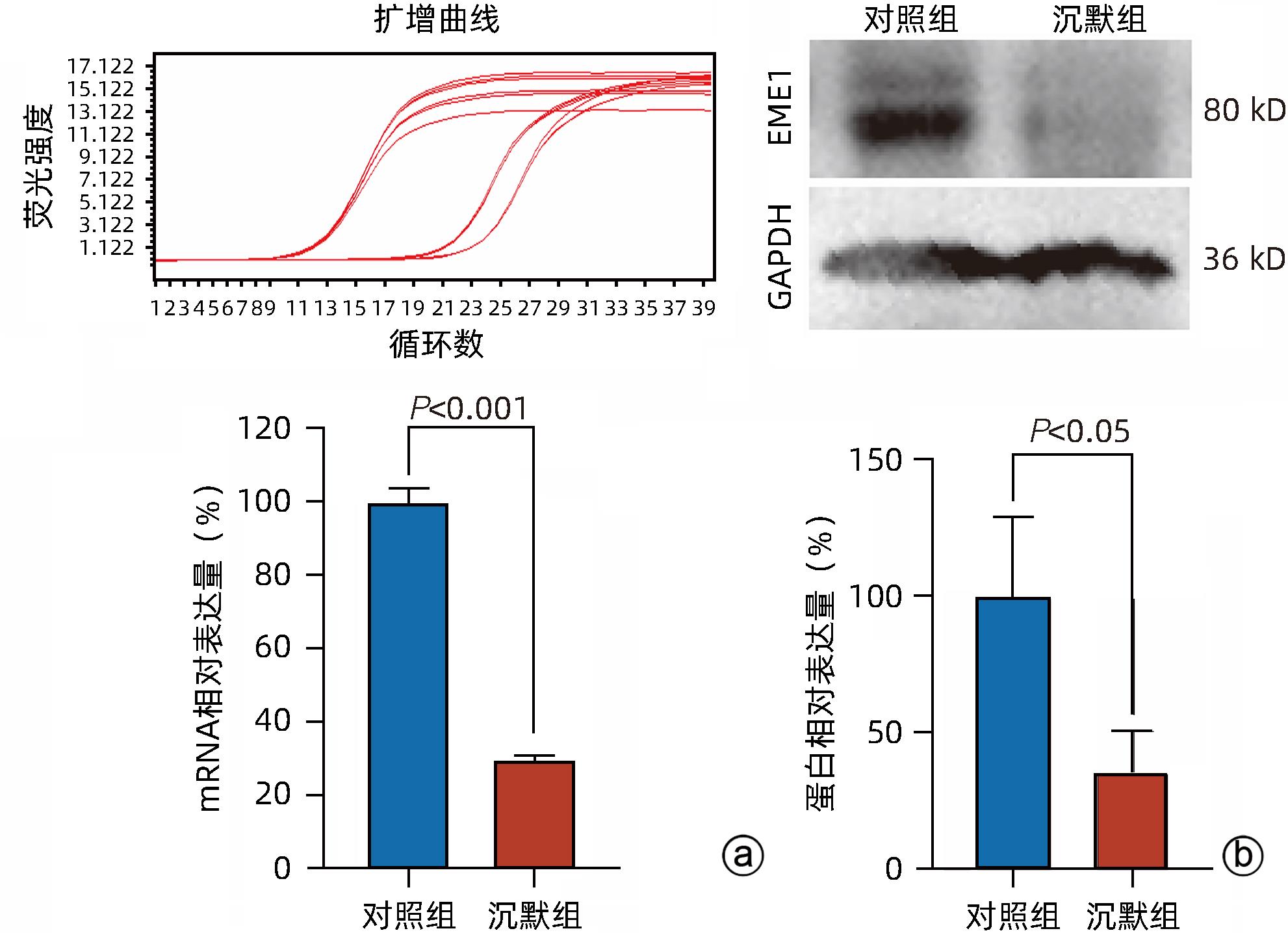
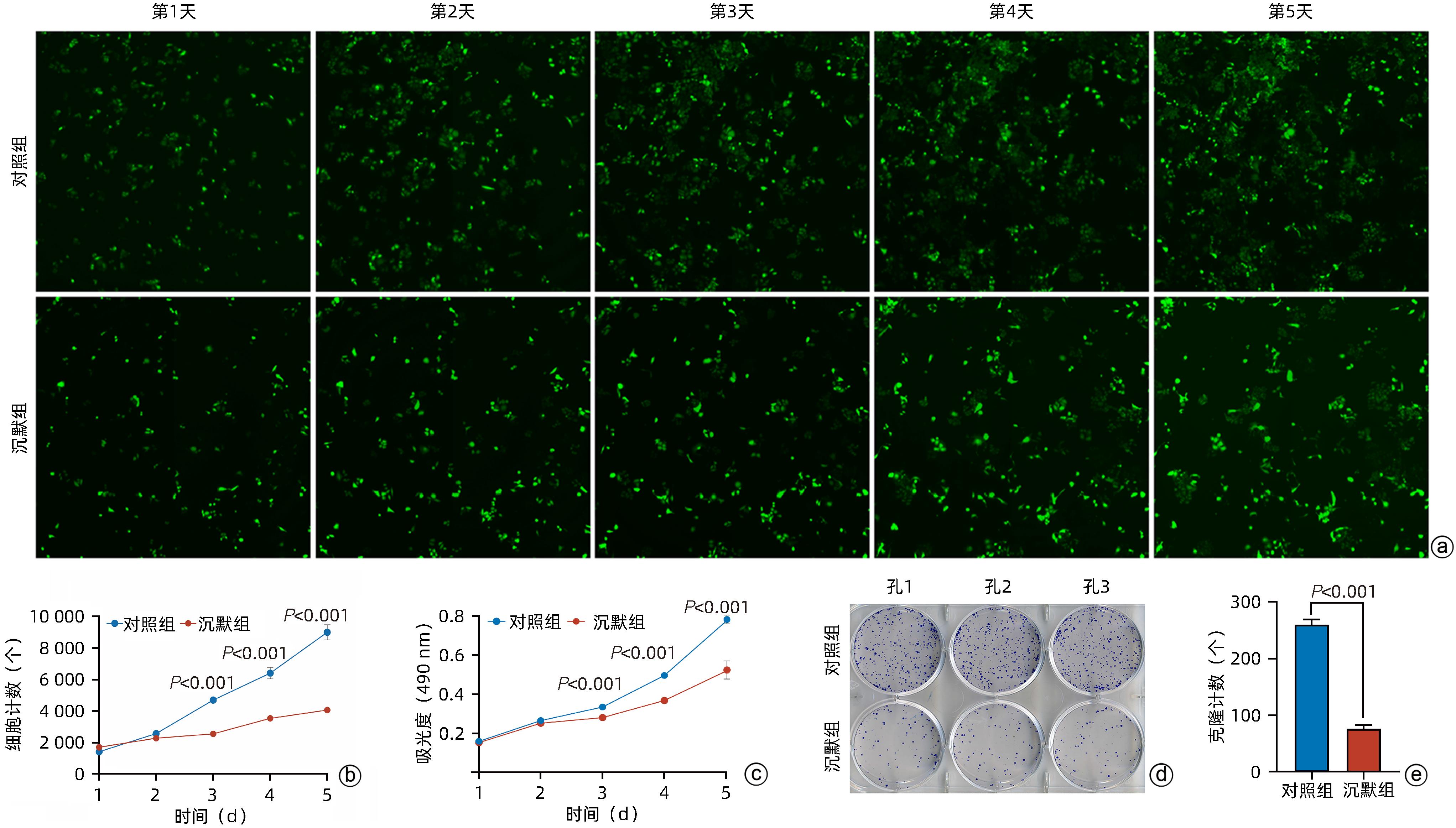
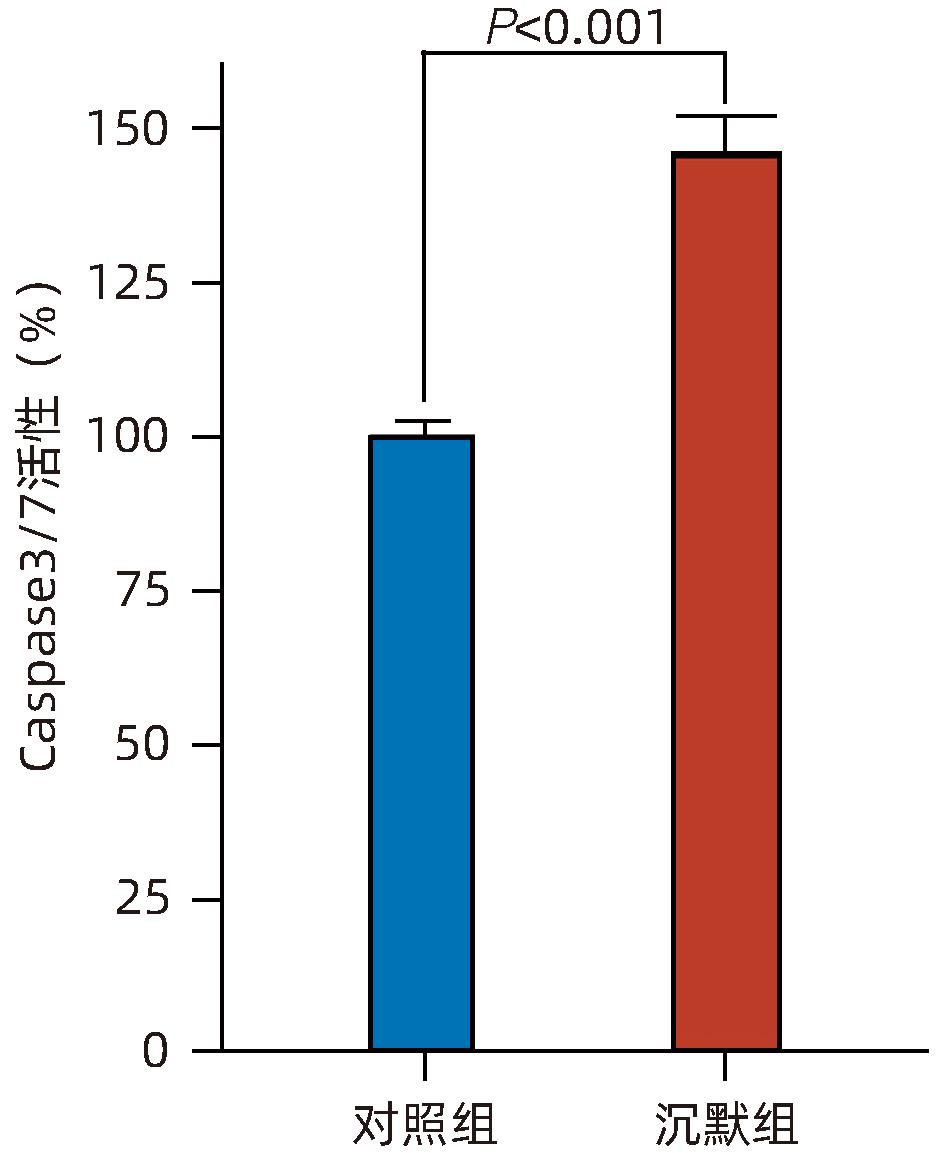
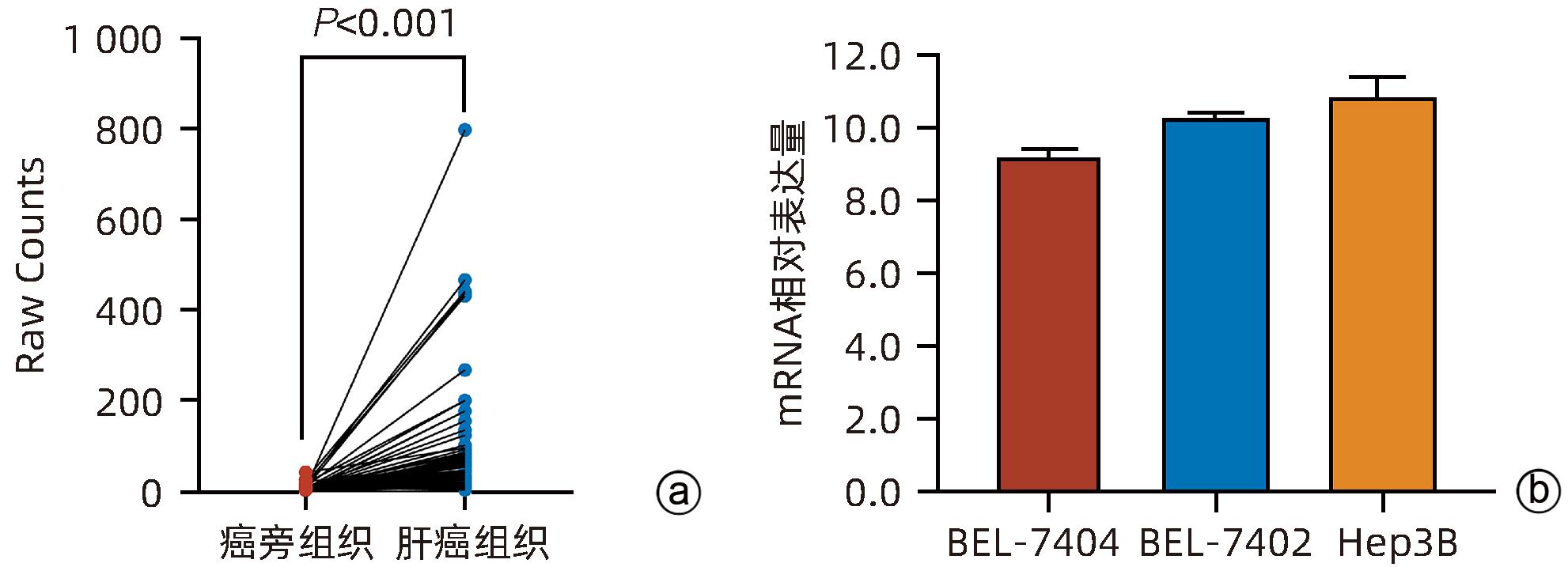
目的 探讨减数分裂内切酶1(EME1)在肝癌组织中的表达及其对肝癌细胞生物学行为的影响。 方法 筛选TCGA数据库肝癌样本中的差异表达基因。采用免疫组化和Western Blot分析EME1在肝癌组织中的表达丰度。通过短发夹RNA(shRNA)构建慢病毒并感染BEL-7404细胞干扰EME1基因表达,分为沉默组(shEME1)和对照组(shCtrl)。通过实时荧光定量PCR法和Western Blot检测两组EME1 mRNA和蛋白表达水平,Celigo计数法及MTT活性检测细胞增殖率,流式细胞术检测细胞周期,Caspase3/7活性检测细胞凋亡。两组间比较采用成组t检验。 结果 TCGA结果显示EME1的mRNA表达水平在肝癌组织中是癌旁组织的18.9倍(114.5±153.0 vs 8.0±7.2,t=5.00,P<0.001);EME1的蛋白表达水平在肝癌组织中是癌旁组织的7.0倍(免疫组化检测,8.4±2.6 vs 1.2±0.4,t=7.55,P<0.001)和2.5倍(Western Blot检测,249.0%±35.5% vs 100.0%±77.8%,t=3.02,P<0.05)。慢病毒感染后,相对于对照组,沉默组EME1的mRNA表达水平下降了29.9%(29.9%± 0.9% vs 100.0%±3.6%,t=32.82,P<0.001),蛋白表达水平显著下降了35.7%(35.7%±14.9% vs 100.0%±28.9%,t=3.42,P<0.05);细胞计数下降了45.1%(4 053±167 vs 8 988±477,t=16.91,P<0.001)、细胞活性下降至66.9%(0.518±0.046 vs 0.774±0.022,t=8.74,P<0.001)及细胞克隆形成能力下降至29.0%(75±6 vs 260±9,t=28.92,P<0.001)。与对照组比较,沉默组G1期细胞(49.9% vs 44.0%,t=8.96,P<0.001)比例增多,G2/M期(15.9% vs 17.9%,t=9.13,P<0.001)与S期(34.2% vs 38.1%,t=6.91,P<0.001)的细胞比例减少;Caspase3/7活性增强了1.5倍(145.8%±5.9% vs 100.0%±2.3%,t=12.50,P<0.001)。 结论 EME1在肝癌组织中高表达,沉默EME1基因可抑制肝癌细胞增殖,促进细胞凋亡。 -
关键词:
- 癌, 肝细胞 /
- 减数分裂内切酶1 /
- RNA, 小分子干扰 /
- 细胞增殖
Abstract:Objective To investigate the expression of essential meiotic endonuclease 1 (EME1) in liver cancer tissue and its effect on the biological behavior of hepatoma cells. Methods The TCGA database was used to identify the differentially expressed genes between liver cancer tissue and paracancerous tissue. Immunohistochemistry and Western Blot were used to measure the expression abundance of EME1 in liver cancer tissue. A lentivirus was constructed by short hairpin RNA, and BEL-7404 cells were transfected with the lentivirus to interfere with the expression of the EME1 gene; the cells were divided into silencing group (shEME1 group) and control group (shCtrl group). Quantitative real-time PCR and Western Blot were used to measure the mRNA and protein expression levels of EME1; Celigo Image Cytometer and MTT assay were used to measure cell proliferation rate; flow cytometry was used to observe cell cycle; Caspase 3/7 activity was used to measure cell apoptosis. The independent-samples t-test was used for comparison between two groups. Results TCGA results showed that the mRNA expression level of EME1 in liver cancer tissue was 18.9 times that in paracancerous tissue (t=5.00, P<0.001), and the protein expression level of EME1 in liver cancer tissue was 7.0 times (based on immunohistochemistry: 8.4±2.6 vs 1.2±0.4, t=7.55, P<0.001) or 2.5 times (based on Western Blot: 249.0%±35.5% vs 100.0%±77.8%, t=3.02, P<0.05) that in paracancerous tissue. After lentivirus infection, compared with the shCtrl group, the shEME1 group had an mRNA expression level of EME1 reduced by 29.9% (29.9%±0.9% vs 100.0%±3.6%, t=32.82, P<0.001), a protein expression level of EME1 reduced by 35.7% (35.7%±14.9% vs 100.0%±28.9%, t=3.42, P<0.05), and a level of cell counting reduced by 45.1% (4 053±167 vs 8 988±477, t=16.91, P<0.001), as well as a level of cell activity reduced to 66.9% (0.518±0.046 vs 0.774±0.022, t=8.74, P<0.001) and a level of colony forming ability reduced to 29.0% (75±6 vs 260±9, t=28.92, P<0.001). Compared with the shCtrl group, the shEME1 group had a significant increase in the proportion of cells in G1 phase (49.9% vs 44.0%, t=8.96, P<0.001) and significant reductions in the proportion of cells in G2/M phase (15.9% vs 17.9%, t=9.13, P<0.001) and S phase (34.2% vs 38.1%, t=6.91, P<0.001), while Caspase 3/7 activity was enhanced by 1.5 times (145.8%±5.9% vs 100.0%±2.3%, t=12.50, P<0.001). Conclusion EME1 is highly expressed in liver cancer tissue, and silencing the EME1 gene can inhibit the proliferation of hepatoma cells and promote cell apoptosis. -
表 1 EME1 shRNA引物序列
Table 1. Sequences of essential meiotic structure-specific endonuclease 1 shRNA
名称 序列(5′-3′) shRNA 正义链:CCGGCCCTGAGAAGACAGGAAAGAAC TCGAGTTCTTTCCTGTCTTCTCAGGGTTTTTG 反义链:AATTCAAAAACCCTGAGAAGACAGGAAAG AACTCGAGTTCTTTCCTGTCTTCTCAGGG shRNA-NC 正义链:CCGGTTCTCCGAACGTGTCACGTTTCA AGAGAAGTGACACGTTCGGAGAATTTTTG 反义链:AATTCAAAAATTCTCCGAACGTGTCAC GTTCTCTT GAAACGTGACACGTTCGGAGAA 鉴定引物 正义链:CCTATTTCCCATGATTCCTTCATA 反义链:GTAATACGGTTATCCACG 表 2 各基因引物序列
Table 2. primer sequences
名称 序列(5′-3′) EME1 正义链: TGACTTCAACAGCGACACCCA 反义链: CACCCTGTTGCTGTAGCCAAA GAPDH 正义链: TCTGAGGAGTTGCCAACATTTG 反义链: GGCTTCACAATCTGAGATGTCAA -
[1] SUNG H, FERLAY J, SIEGEL RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71( 3): 209- 249. DOI: 10.3322/caac.21660. [2] LLOVET JM, KELLEY RK, VILLANUEVA A, et al. Hepatocellular carcinoma[J]. Nat Rev Dis Primers, 2021, 7: 6. DOI: 10.1038/s41572-020-00240-3. [3] CHEN SH, WANG CY, GUO C, et al. Establishment of a nomogram model for predicting the survival of hepatitis B virus-related hepatocellular carcinoma[J]. J Clin Hepatol, 2022, 38( 7): 1566- 1571. DOI: 10.3969/j.issn.1001-5256.2022.07.020.陈松海, 王春艳, 郭畅, 等. 预测HBV相关肝细胞癌生存的列线图模型的建立[J]. 临床肝胆病杂志, 2022, 38( 7): 1566- 1571. DOI: 10.3969/j.issn.1001-5256.2022.07.020. [4] WANG JJ, WANG KX, CHEN C, et al. Survival analysis and development of a prognostic nomogram for patients with hepatitis B virus-associated hepatocellular carcinoma[J]. Heliyon, 2023, 9( 10): e20850. DOI: 10.1016/j.heliyon.2023.e20850. [5] JI D, CHEN Y, BI JF, et al. Entecavir plus Biejia-Ruangan compound reduces the risk of hepatocellular carcinoma in Chinese patients with chronic hepatitis B[J]. J Hepatol, 2022, 77( 6): 1515- 1524. DOI: 10.1016/j.jhep.2022.07.018. [6] GWON GH, JO A, BAEK K, et al. Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates[J]. EMBO J, 2014, 33( 9): 1061- 1072. DOI: 10.1002/embj.201487820. [7] HAN YY, ZHENG QY, TIAN Y, et al. Identification of a nine-gene panel as a prognostic indicator for recurrence with muscle-invasive bladder cancer[J]. J Surg Oncol, 2019, 119( 8): 1145- 1154. DOI: 10.1002/jso.25446. [8] VÉQUAUD E, DESPLANQUES G, JÉZÉQUEL P, et al. Survivin contributes to DNA repair by homologous recombination in breast cancer cells[J]. Breast Cancer Res Treat, 2016, 155( 1): 53- 63. DOI: 10.1007/s10549-015-3657-z. [9] PENG L, LIANG J, WANG Q, et al. A DNA damage repair gene signature associated with immunotherapy response and clinical prognosis in clear cell renal cell carcinoma[J]. Front Genet, 2022, 13: 798846. DOI: 10.3389/fgene.2022.798846. [10] GUO ZG, LIANG EB, LI W, et al. Essential meiotic structure-specific endonuclease1(EME1) promotes malignant features in gastric cancer cells via the Akt/GSK3B/CCND1 pathway[J]. Bioengineered, 2021, 12( 2): 9869- 9884. DOI: 10.1080/21655979.2021.1999371. [11] WANG YX, HUANG XL, SU ZH, et al. The Glu69Asp polymorphism of EME1 gene is associated with an increased risk of hepatocellular carcinoma in Guangxi population, China[J]. Int J Gen Med, 2022, 15: 7855- 7866. DOI: 10.2147/IJGM.S383261. [12] ZHAO JW, LIU L, ZHANG AQ, et al. Effect of EME1 exon variant Ile350Thr on risk and early onset of breast cancer in southern Chinese women[J]. J Biomed Res, 2013, 27( 3): 193- 201. DOI: 10.7555/JBR.27.20130013. [13] SUZUKI T, YASUI M, HONMA M. Mutator phenotype and DNA double-strand break repair in BLM helicase-deficient human cells[J]. Mol Cell Biol, 2016, 36( 23): 2877- 2889. DOI: 10.1128/MCB.00443-16. [14] SHAMMAS MA, SHMOOKLER REIS RJ, KOLEY H, et al. Dysfunctional homologous recombination mediates genomic instability and progression in myeloma[J]. Blood, 2009, 113( 10): 2290- 2297. DOI: 10.1182/blood-2007-05-089193. [15] FULDA S. Tumor resistance to apoptosis[J]. Int J Cancer, 2009, 124( 3): 511- 515. DOI: 10.1002/ijc.24064. [16] SZAKAL B, BRANZEI D. Premature Cdk1/Cdc5/Mus81 pathway activation induces aberrant replication and deleterious crossover[J]. EMBO J, 2013, 32( 8): 1155- 1167. DOI: 10.1038/emboj.2013.67. [17] PALMA A, PUGLIESE GM, MURFUNI I, et al. Phosphorylation by CK2 regulates MUS81/EME1 in mitosis and after replication stress[J]. Nucleic Acids Res, 2018, 46( 10): 5109- 5124. DOI: 10.1093/nar/gky280. [18] GUO YG, ZHU ZH, HUANG ZY, et al. CK2-induced cooperation of HHEX with the YAP-TEAD4 complex promotes colorectal tumorigenesis[J]. Nat Commun, 2022, 13( 1): 4995. DOI: 10.1038/s41467-022-32674-6. [19] KWON J, ZHANG JM, MOK B, et al. CK2-mediated phosphorylation upregulates the stability of USP13 and promotes ovarian cancer cell proliferation[J]. Cancers, 2022, 15( 1): 200. DOI: 10.3390/cancers15010200. [20] PITRODA SP, PASHTAN IM, LOGAN HL, et al. DNA repair pathway gene expression score correlates with repair proficiency and tumor sensitivity to chemotherapy[J]. Sci Transl Med, 2014, 6( 229): 229ra42. DOI: 10.1126/scitranslmed.3008291. [21] XU YH, LI J, ZHU KK, et al. FIBP interacts with transcription factor STAT3 to induce EME1 expression and drive radioresistance in lung adenocarcinoma[J]. Int J Biol Sci, 2023, 19( 12): 3816- 3829. DOI: 10.7150/ijbs.83134. [22] WEINANDY A, PIROTH MD, GOSWAMI A, et al. Cetuximab induces eme1-mediated DNA repair: A novel mechanism for cetuximab resistance[J]. Neoplasia, 2014, 16( 3): 207- 220, 220.e1- 220. e4. DOI: 10.1016/j.neo.2014.03.004. -



 PDF下载 ( 1933 KB)
PDF下载 ( 1933 KB)

 下载:
下载: