microRNA-933对LX-2细胞凋亡和增殖的影响及其分子机制
DOI: 10.12449/JCH240716
利益冲突声明:本文不存在任何利益冲突。
作者贡献声明:海龙、丁向春负责课题设计,资料分析,撰写论文;海龙、马丽娜、雒夏参与收集数据,修改论文;丁向春负责拟定写作思路,指导撰写文章并最后定稿。
Effect of miRNA-933 on the apoptosis and proliferation of LX-2 cells and its molecular mechanism
-
摘要:
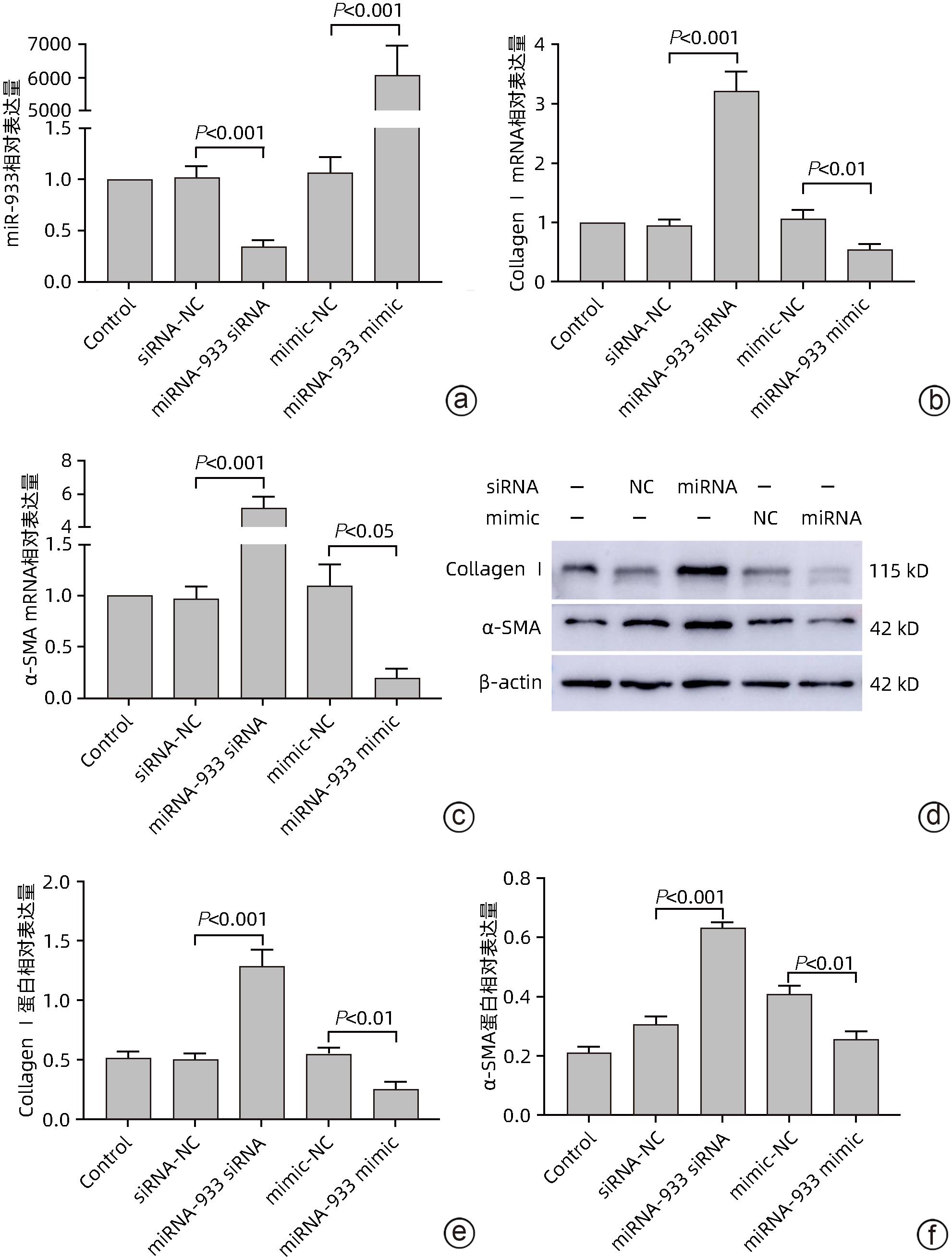
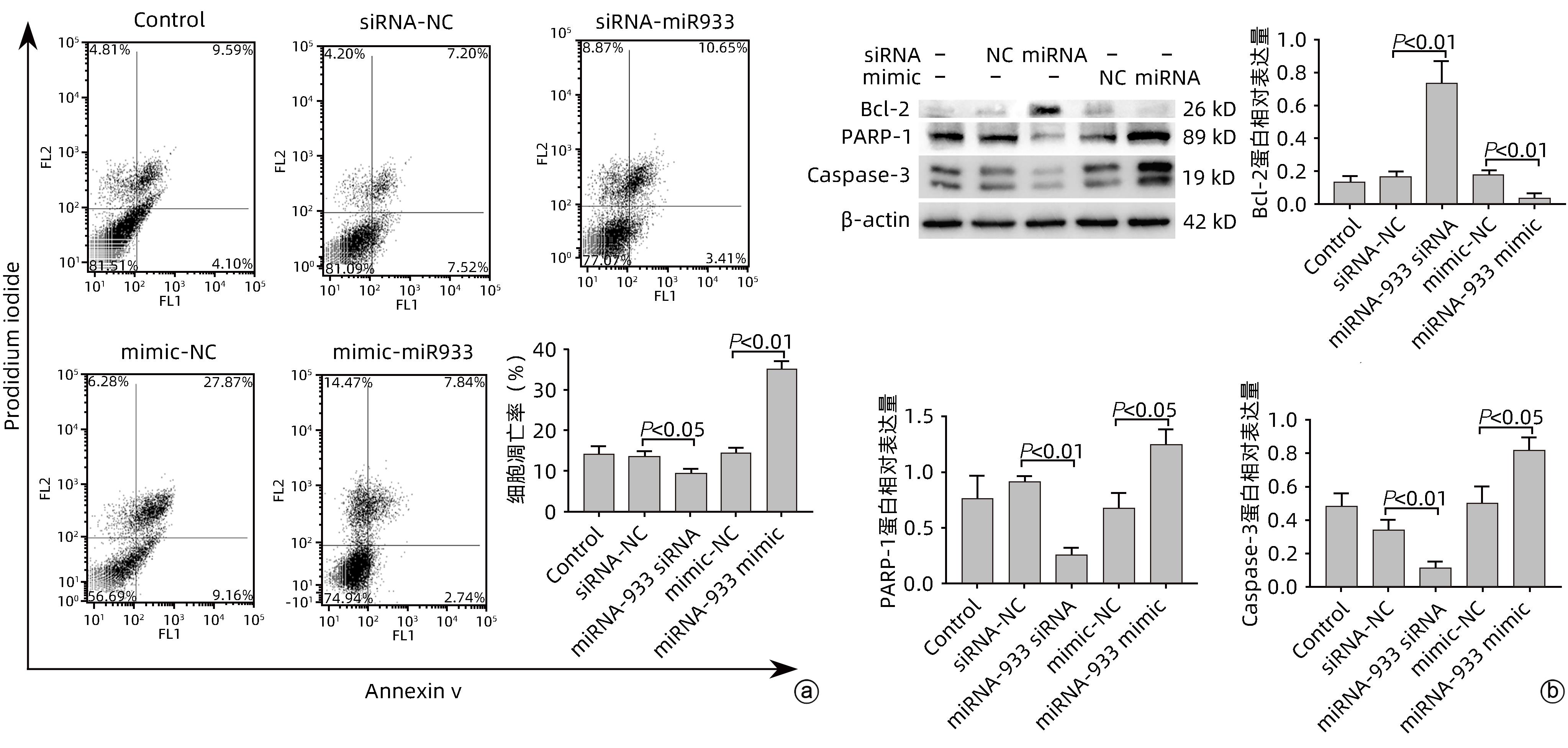
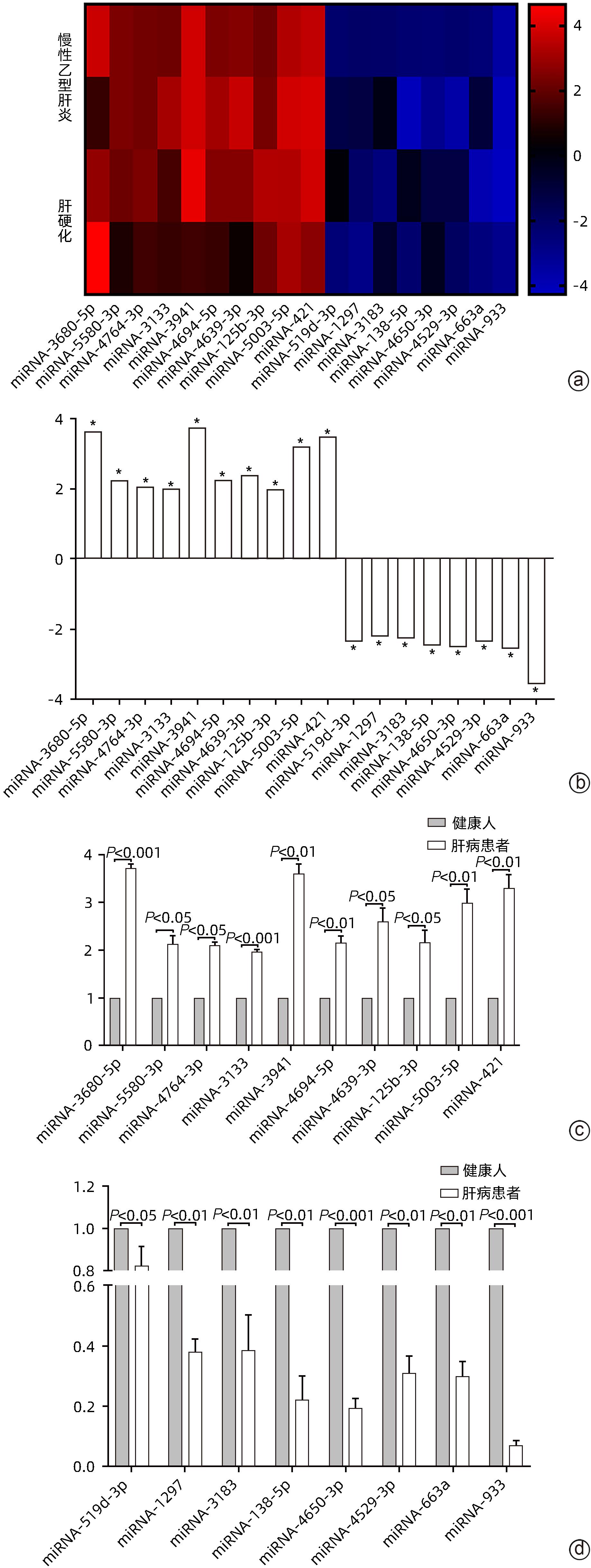
目的 探讨microRNA-933(miRNA-933)对人肝星状细胞系LX-2细胞凋亡和增殖的调控作用及其机制。 方法 首先以人肝组织为研究对象,利用基因芯片技术,检测肝硬化与慢性乙型肝炎肝组织相较于正常肝组织中差异表达的基因,并从中筛选差异表达显著的miRNA,从而确定研究对象为miRNA-933。进而以人肝星状细胞系LX-2为研究对象,利用miRNA-933分子模拟剂(miRNA-933 mimic)与抑制剂(miRNA-933 siRNA),构建LX-2过表达与敲减模型,并以转染mimic-NC(过表达)或siRNA-NC(敲减)的细胞作为阴性对照。使用实时荧光定量PCR(qPCR)和Western Blot技术,检测miRNA-933与活化标志蛋白的表达水平;随后采用细胞增殖实验、流式细胞术等技术,检测miRNA-933对细胞凋亡、增殖和活化的影响,并探讨其机制。计量资料两组间比较采用成组t检验,多组间比较采用单因素方差分析,并经过Bonferroni校正。 结果 基于基因芯片检测结果,最终筛选出18个显著差异表达的miRNA,其中miRNA-933表达显著下调(P<0.05)。miRNA-933 mimic与siRNA转染LX-2细胞后,结果显示,与阴性对照组相比,miRNA-933 siRNA显著下调miRNA-933的表达(P=0.000 7),而miRNA-933 mimic显著上调miRNA-933的表达(P=0.000 3);Western Blot和qPCR检测结果表明,miRNA-933 siRNA显著上调胶原蛋白Ⅰ(Collagen Ⅰ)和α-平滑肌肌动蛋白(α-SMA)的表达(P值均<0.001),而miRNA-933 mimic显著抑制Collagen Ⅰ和α-SMA的表达(P值均<0.05)。流式细胞术检测结果显示,与阴性对照组相比,miRNA-933 siRNA显著下调LX-2细胞凋亡率(P=0.031 9),miRNA-933 mimic显著上调LX-2细胞凋亡率(P=0.005 5);Western Blot检测结果表明,与阴性对照组相比,miRNA-933 siRNA可以抑制LX-2细胞中胱天蛋白酶3(Caspase-3)和多聚ADP核糖聚合酶1(PARP-1)的表达(P值分别为0.006 7、0.003 0),上调B淋巴细胞瘤-2(Bcl-2)的表达(P=0.002 0),而miRNA-933 mimic可显著上调Caspase-3(P=0.011 8)和PARP-1(P=0.049 5)的表达,下调Bcl-2的表达(P=0.002 1)。细胞增殖实验结果显示,与阴性对照组相比,miRNA-933 siRNA可促进LX-2细胞增殖(P=0.011 5);相反miRNA-933 mimic可抑制LX-2细胞增殖(P=0.001 2)。Western Blot和qPCR检测显示,miRNA-933 siRNA显著抑制Kruppel样因子6(KLF6)的表达,进而下调活化转录因子4(ATF4)、活化转录因子3(ATF3)、C/EBP同源蛋白(CHOP)蛋白的表达,而miRNA-933 mimic会促进以上蛋白的表达(P值均<0.05)。 结论 miRNA-933可能通过促进LX-2细胞中KLF6/ATF4/ATF3/CHOP/Bcl-2信号轴的激活,进而促进细胞凋亡,抑制细胞的活化和增殖。 Abstract:Objective To investigate the regulatory effect of miRNA-933 on the apoptosis and proliferation of human hepatic stellate cell line LX-2 and its mechanism. Methods Firstly, with human liver tissue for research, gene microarray technology was used to detect the differentially expressed genes in liver tissue between liver cirrhosis/chronic hepatitis B tissue and normal liver tissue, among which the significantly differentially expressed miRNAs were identified, and thus miRNA-933 was determined as the research object. Then, with the human hepatic stellate cell line LX-2 for research, miRNA-933 mimic and inhibitor (miRNA-933 siRNA) were used to construct the LX-2 models of overexpression and knockdown, and the cells transfected with mimic-NC (overexpression) or siRNA-NC (knockdown) were established as the negative control group. Quantitative real-time PCR and Western blot were used to measure the expression levels of miRNA-933 and activation biomarkers; techniques such as cell proliferation assay and flow cytometry were used to investigate the effect and mechanism of miRNA-933 on cell apoptosis, proliferation, and activation. The independent-samples t test was used for comparison of continuous data between two groups; a one-way analysis of variance was used for comparison between multiple groups, and Bonferroni correction was also performed. Results A total of 18 significantly differentially expressed miRNAs were obtained based on the results of gene microarray, among which miRNA-933 was significantly downregulated (P<0.05). After LX-2 cells were transfected with miRNA-933 mimic or siRNA, compared with the negative control group, miRNA-933 siRNA significantly downregulated the expression of miRNA-933 (P=0.000 7), while miRNA-933 mimic significantly upregulated the expression of miRNA-933 (P=0.000 3). Western blot and quantitative real-time PCR showed that miRNA-933 siRNA significantly upregulated the expression of collagen I and α-SMA (P<0.001), while miRNA-933 mimic significantly inhibited the expression of collagen I and α-SMA (P<0.05). Flow cytometry showed that compared with the negative control group, miRNA-933 siRNA significantly downregulated the apoptosis rate of LX-2 cells (P=0.031 9), and miRNA-933 mimic significantly upregulated the apoptosis rate of LX-2 cells (P=0.005 5). Western blot showed that compared with the negative control group, miRNA-933 siRNA could inhibit the expression of Caspase-3 (P=0.006 7) and poly(ADP-ribose) polymerase-1 (PARP-1) (P=0.003 0) and upregulate the expression of B-cell lymphoma-2 (Bcl-2) in LX-2 cells (P=0.002 0), while miRNA-933 mimic could significantly upregulate the expression of Caspase-3 (P=0.011 8) and PARP-1 (P=0.049 5) and downregulated the expression of Bcl-2 (P=0.002 1). Cell proliferation assay showed that compared with the negative control group, miRNA-933 siRNA could promote the proliferation of LX-2 cells (P=0.011 5), while on the contrary, miRNA-933 mimic could inhibit the proliferation of LX-2 cells (P=0.001 2). Western blot and quantitative real-time PCR showed that miRNA-933 siRNA significantly inhibited the expression of Kruppel-like factor 6 (KLF6) and downregulated the expression of activating transcription factor 4 (ATF4), activating transcription factor 3 (ATF3), and C/EBP homologous protein (CHOP), while miRNA-933 mimic promoted the expression of the above proteins (all P<0.05). Conclusion This study shows that miRNA-933 may promote cell apoptosis and inhibit cell activation and proliferation by promoting the activation of the KLF6/ATF4/ATF3/CHOP/Bcl-2 signal axis in LX-2 cells. -
Key words:
- Hepatic Fibrosis /
- MicroRNAs /
- Hepatic Stellate Cells
-
表 1 引物序列
Table 1. Primer sequences
引物 序列(5′-3′) miRNA-933 F:ACACTCCAGCTGGCTGTGTGCGCAGGG‑AGACCTC R:TGGTGTCGTGGAGTCG U6 F:CTCGCTTCGGCAGCACA R:AACGCTTCACGAATTTGCGT miRNA-933-RT UGUGCGCAGGGAGACCUCUCCC miRNA-933 mimic CAAAGUGCCUCCCUUUAGAGUG NC mimic UUCUCCGAACGUGUCACGUTT miRNA-933 siRNA GUUUCACGGAGGGAAAUCUCAC NC siRNA CAGUACUUUUGUGUAGUACCAA -
[1] TSUCHIDA T, FRIEDMAN SL. Mechanisms of hepatic stellate cell activation[J]. Nat Rev Gastroenterol Hepatol, 2017, 14( 7): 397- 411. DOI: 10.1038/nrgastro.2017.38. [2] ZHANG CY, YUAN WG, HE P, et al. Liver fibrosis and hepatic stellate cells: Etiology, pathological hallmarks and therapeutic targets[J]. World J Gastroenterol, 2016, 22( 48): 10512- 10522. DOI: 10.3748/wjg.v22.i48.10512. [3] HIGASHI T, FRIEDMAN SL, HOSHIDA Y. Hepatic stellate cells as key target in liver fibrosis[J]. Adv Drug Deliv Rev, 2017, 121: 27- 42. DOI: 10.1016/j.addr.2017.05.007. [4] MARTÍ-RODRIGO A, ALEGRE F, ÁB MORAGREGA, et al. Rilpivirine attenuates liver fibrosis through selective STAT1-mediated apoptosis in hepatic stellate cells[J]. Gut, 2020, 69( 5): 920- 932. DOI: 10.1136/gutjnl-2019-318372. [5] RUPAIMOOLE R, SLACK FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases[J]. Nat Rev Drug Discov, 2017, 16( 3): 203- 222. DOI: 10.1038/nrd.2016.246. [6] JIANG XP, AI WB, WAN LY, et al. The roles of microRNA families in hepatic fibrosis[J]. Cell Biosci, 2017, 7: 34. DOI: 10.1186/s13578-017-0161-7. [7] LIN HY, WANG FS, YANG YL, et al. MicroRNA-29a suppresses CD36 to ameliorate high fat diet-induced steatohepatitis and liver fibrosis in mice[J]. Cells, 2019, 8( 10): 1298. DOI: 10.3390/cells8101298. [8] SONG L, CHEN TY, ZHAO XJ, et al. Pterostilbene prevents hepatocyte epithelial-mesenchymal transition in fructose-induced liver fibrosis through suppressing miR-34a/Sirt1/p53 and TGF-β1/Smads signalling[J]. Br J Pharmacol, 2019, 176( 11): 1619- 1634. DOI: 10.1111/bph.14573. [9] LU TX, ROTHENBERG ME. MicroRNA[J]. J Allergy Clin Immunol, 2018, 141( 4): 1202- 1207. DOI: 10.1016/j.jaci.2017.08.034. [10] LOU WY, LIU JX, GAO YJ, et al. MicroRNA regulation of liver cancer stem cells[J]. Am J Cancer Res, 2018, 8( 7): 1126- 1141. [11] ROEHLEN N, CROUCHET E, BAUMERT TF. Liver fibrosis: Mechanistic concepts and therapeutic perspectives[J]. Cells, 2020, 9( 4): 875. DOI: 10.3390/cells9040875. [12] EZHILARASAN D, SOKAL E, NAJIMI M. Hepatic fibrosis: It is time to go with hepatic stellate cell-specific therapeutic targets[J]. Hepatobiliary Pancreat Dis Int, 2018, 17( 3): 192- 197. DOI: 10.1016/j.hbpd.2018.04.003. [13] LI J, CHAN MC, YU Y, et al. MiR-29b contributes to multiple types of muscle atrophy[J]. Nat Commun, 2017, 8: 15201. DOI: 10.1038/ncomms15201. [14] CAI FF, BIAN YQ, WU R, et al. Yinchenhao decoction suppresses rat liver fibrosis involved in an apoptosis regulation mechanism based on network pharmacology and transcriptomic analysis[J]. Biomed Pharmacother, 2019, 114: 108863. DOI: 10.1016/j.biopha.2019.108863. [15] HUANG YH, TIAO MM, HUANG LT, et al. Activation of miR-29a in activated hepatic stellate cells modulates its profibrogenic phenotype through inhibition of histone deacetylases 4[J]. PLoS One, 2015, 10( 8): e0136453. DOI: 10.1371/journal.pone.0136453. [16] GUO CJ, PAN Q, LI DG, et al. MiR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: An essential role for apoptosis[J]. J Hepatol, 2009, 50( 4): 766- 778. DOI: 10.1016/j.jhep.2008.11.025. [17] ZHU XQ, SHAO P, TANG YC, et al. hsa_circRNA_100533 regulates GNAS by sponging hsa_miR_933 to prevent oral squamous cell carcinoma[J]. J Cell Biochem, 2019, 120( 11): 19159- 19171. DOI: 10.1002/jcb.29245. [18] LI HZ, ZHANG YW, XU YJ, et al. MiR-933 inhibits proliferation, migration and invasion of lung cancer cell lines by regulation of KLF6 gene[J]. China Oncol, 2021, 31( 7): 581- 588. DOI: 10.19401/j.cnki.1007-3639.2021.07.004.李海洲, 张艳炜, 许英杰, 等. miR-933调控KLF6基因影响非小细胞肺癌的作用研究[J]. 中国癌症杂志, 2021, 31( 7): 581- 588. DOI: 10.19401/j.cnki.1007-3639.2021.07.004. [19] ZHANG YT, MA YY, XU WH, et al. Association of microRNA-933 variant with the susceptibility to gastric cancer[J]. J BUON, 2017, 22( 2): 390- 395. [20] ZHOU B, GUO H, TANG J. Long non-coding RNA TFAP2A-AS1 inhibits cell proliferation and invasion in breast cancer via miR-933/SMAD2[J]. Med Sci Monit, 2019, 25: 1242- 1253. DOI: 10.12659/MSM.912421. [21] CHENG Y, SHI WW, CUI XD, et al. Long noncoding RNA TFAP2A-AS1 suppressed hepatitis B virus replication by modulating miR-933/HDAC11[J]. Dis Markers, 2022, 2022: 7733390. DOI: 10.1155/2022/7733390. [22] ISLAM ABMMK, MOHAMMAD E, KHAN MA. Aberration of the modulatory functions of intronic microRNA hsa-miR-933 on its host gene ATF2 results in type II diabetes mellitus and neurodegenerative disease development[J]. Hum Genomics, 2020, 14( 1): 34. DOI: 10.1186/s40246-020-00285-1. [23] RANE MJ, ZHAO YG, CAI L. Krϋppel-like factors(KLFs) in renal physiology and disease[J]. EBioMedicine, 2019, 40: 743- 750. DOI: 10.1016/j.ebiom.2019.01.021. [24] SYAFRUDDIN SE, RODRIGUES P, VOJTASOVA E, et al. A KLF6-driven transcriptional network links lipid homeostasis and tumour growth in renal carcinoma[J]. Nat Commun, 2019, 10( 1): 1152. DOI: 10.1038/s41467-019-09116-x. [25] LANG UE, KOCABAYOGLU P, CHENG GZ, et al. GSK3β phosphorylation of the KLF6 tumor suppressor promotes its transactivation of p21[J]. Oncogene, 2013, 32( 38): 4557- 4564. DOI: 10.1038/onc.2012.457. [26] CAI MG, SHAO W, YU HJ, et al. Paeonol inhibits cell proliferation, migration and invasion and induces apoptosis in hepatocellular carcinoma by regulating miR-21-5p/KLF6 axis[J]. Cancer Manag Res, 2020, 12: 5931- 5943. DOI: 10.2147/CMAR.S254485. [27] ZHANG N, YAN QQ, LU L, et al. The KLF6 splice variant KLF6-SV1 promotes proliferation and invasion of non-small cell lung cancer by up-regultating PI3K-AKT signaling pathway[J]. J Cancer, 2019, 10( 22): 5324- 5331. DOI: 10.7150/jca.34212. [28] CHIAM K, RYAN NK, RICCIARDELLI C, et al. Characterization of the prostate cancer susceptibility gene KLF6 in human and mouse prostate cancers[J]. Prostate, 2013, 73( 2): 182- 193. DOI: 10.1002/pros.22554. [29] GHIASSI-NEJAD Z, HERNANDEZ-GEA V, WOODRELL C, et al. Reduced hepatic stellate cell expression of Kruppel-like factor 6 tumor suppressor isoforms amplifies fibrosis during acute and chronic rodent liver injury[J]. Hepatology, 2013, 57( 2): 786- 796. DOI: 10.1002/hep.26056. [30] TIAN F, ZHAO JZ, BU SC, et al. KLF6 induces apoptosis in human lens epithelial cells through the ATF4-ATF3-CHOP axis[J]. Drug Des Devel Ther, 2020, 14: 1041- 1055. DOI: 10.2147/DDDT.S218467. -



 PDF下载 ( 1929 KB)
PDF下载 ( 1929 KB)

 下载:
下载: