自噬途径降解肝脏脂滴的研究进展
DOI: 10.12449/JCH240931
Research advances in the degradation of hepatic lipid droplets through the autophagy pathway
-
摘要: 自噬是一种高度保守的细胞降解途径,可通过“脂噬”过程来降解脂滴。脂噬可以选择性地识别脂类物质并将其降解,促进β氧化,进而维持细胞内脂质代谢的平衡状态。肝脏通过脂噬信号通路或关键分子来调控脂滴代谢,进而降低肝脏脂肪变性,改善非酒精性脂肪性肝病。本文总结归纳了巨自噬、分子伴侣介导的自噬和微自噬样3种自噬途径降解肝脏脂滴的最新研究进展,AMPK/mTOR-ULK1、ATGL-SIRT1、FGF21-JMJD3、Akt作为调控脂噬过程的主要信号通路,有助于维持肝脂质代谢稳态,能够为临床预防和治疗非酒精性脂肪性肝病提供新思路。Abstract: Autophagy is a highly conserved cellular degradation pathway that degrades lipid droplets through a process called “lipophagy”. Lipophagy can selectively recognize lipid substances and degrade them, promoting β oxidation and thereby maintaining the balance of intracellular lipid metabolism. The liver regulates lipid droplet metabolism through lipophagy signaling pathways or key molecules, thereby alleviating hepatic steatosis and improving nonalcoholic fatty liver disease (NAFLD). This article reviews the latest advances in the degradation of hepatic lipid droplets through the three autophagic pathways of macroautophagy, molecular chaperone-mediated autophagy, and microautophagy. The major signaling pathways of AMPK/mTOR-ULK1, ATGL-SIRT1, FGF21-JMJD3, and Akt are involved in the regulation of the lipophagy process and help to maintain the homeostasis of lipid metabolism in the liver, so as to provide new ideas for clinical prevention and treatment of NAFLD.
-
Key words:
- Non-alcoholic Fatty Liver Disease /
- Autophagy /
- Lipid Metabolism /
- Pathologic Processes
-
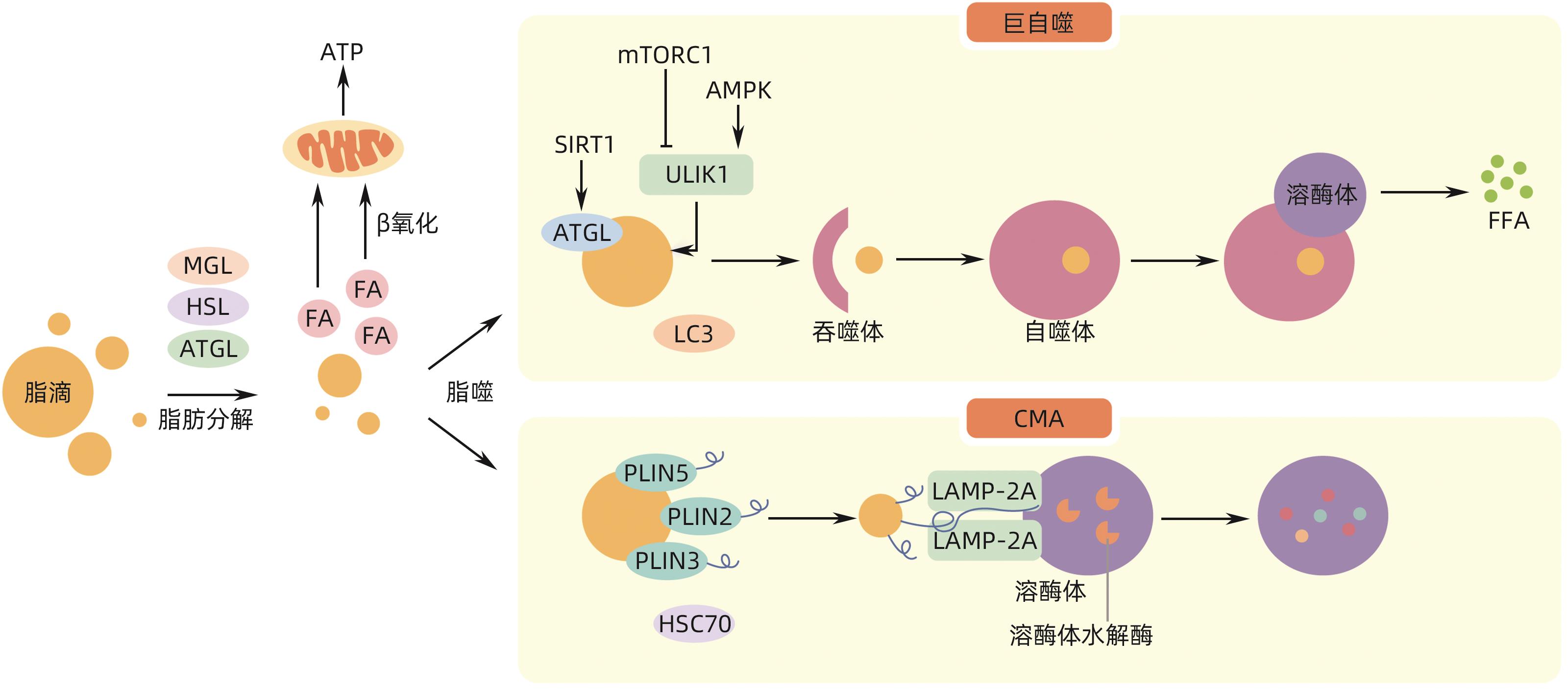
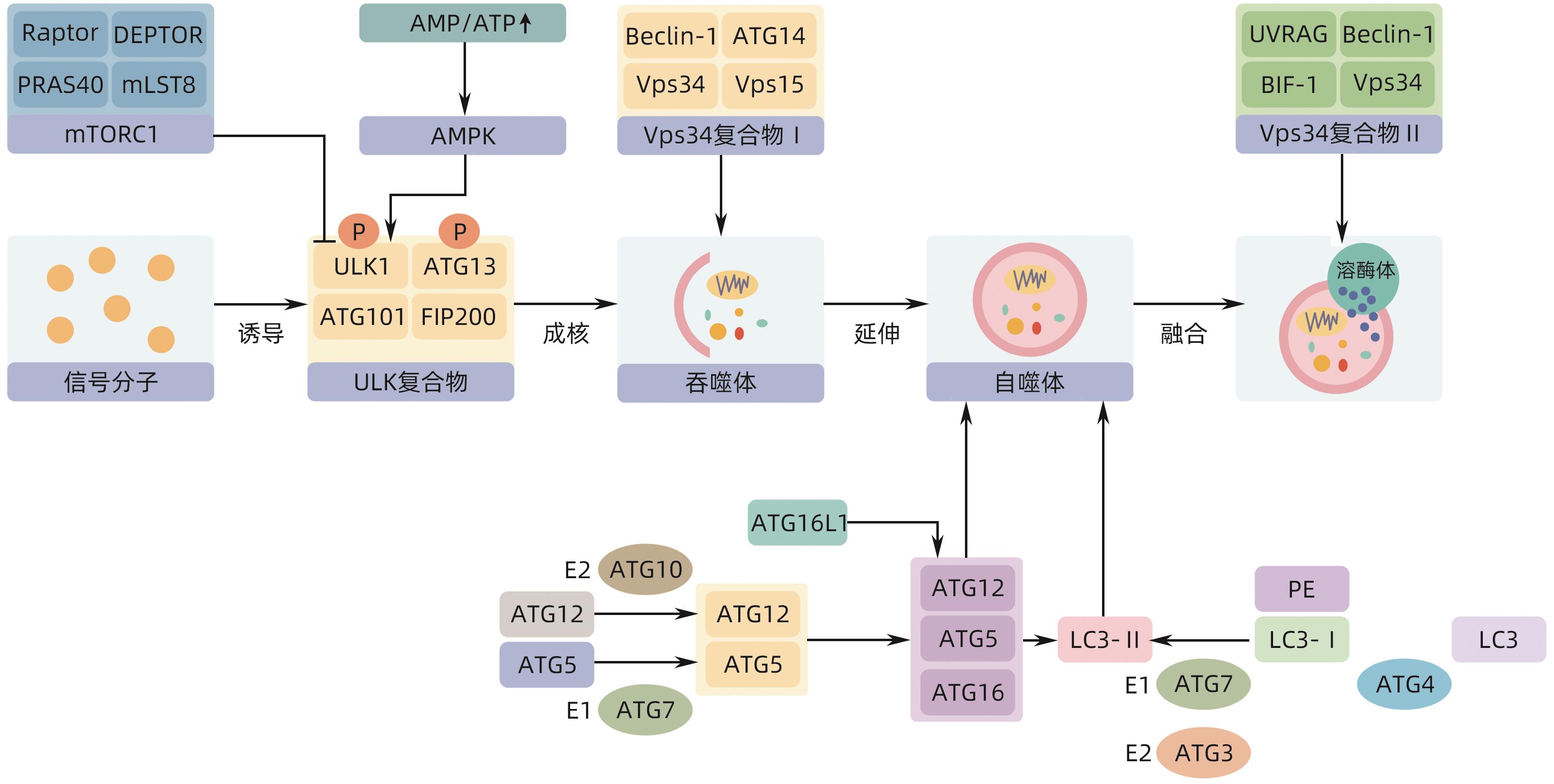
表 1 不同自噬形式参与脂滴降解的分子机制
Table 1. Molecular mechanisms of different forms of autophagy involved in lipid droplet degradation
自噬类型 关键分子机制及通路 作用 生物 文献 巨自噬 ULK1/ATG1 启动自噬体的形成 哺乳动物、酵母 [14] mTOR-ULK1 抑制脂噬 哺乳动物 [15] AMPK-ULK1 促进脂噬 哺乳动物 [15] TFEB 脂噬主要调节因子 哺乳动物 [16] HLH-30、MXL-3 调节脂噬 秀丽隐杆线虫 [16] FgATG15(酵母ATG15同源物) 参与脂滴分解 禾谷镰刀菌 [17] OsATG7(酵母ATG7同源物) 参与脂滴分解 水稻 [18] ATG蛋白 识别脂滴,促进自噬体的形成 酵母、哺乳动物、植物、微藻 [19-21] SNARE蛋白 介导自噬体膜的扩展/闭合 酵母、哺乳动物 [22-23] SQSTM1/p62 桥接脂滴与吞噬泡 哺乳动物 [24] Rab GTP酶 介导脂滴的募集 哺乳动物 [25] CMA HSC70 识别并结合脂滴 哺乳动物 [26] LAMP-2A 结合脂滴并将其移至溶酶体内 哺乳动物 [26] 微自噬 ATG6和ATG14 形成脂滴募集位点 酵母 [27] ESCRT蛋白Vps27 将脂滴转移到液泡 酵母 [28] -
[1] YOUNOSSI ZM, GOLABI P, PAIK JM, et al. The global epidemiology of nonalcoholic fatty liver disease(NAFLD) and nonalcoholic steatohepatitis(NASH): a systematic review[J]. Hepatology, 2023, 77( 4): 1335- 1347. DOI: 10.1097/HEP.0000000000000004. [2] SARIN SK, KUMAR M, ESLAM M, et al. Liver diseases in the Asia-Pacific region: a Lancet Gastroenterology& Hepatology Commission[J]. Lancet Gastroenterol Hepatol, 2020, 5( 2): 167- 228. DOI: 10.1016/S2468-1253(19)30342-5. [3] ANDROUTSAKOS T, NASIRI-ANSARI N, BAKASIS AD, et al. SGLT-2 inhibitors in NAFLD: expanding their role beyond diabetes and cardioprotection[J]. Int J Mol Sci, 2022, 23( 6). DOI: 10.3390/ijms23063107. [4] KOU XX, ZHANG H, DENG JX, et al. Role of intrahepatic microenvironment induced-autophagy in nonalcoholic fatty liver disease[J]. J Clin Hepatol, 2023, 39( 6): 1440- 1445. DOI: 10.3969/j.issn.1001-5256.2023.06.029.寇萱萱, 张华, 邓婧鑫, 等. 肝内微环境诱导的自噬在非酒精性脂肪性肝病中的作用[J]. 临床肝胆病杂志, 2023, 39( 6): 1440- 1445. DOI: 10.3969/j.issn.1001-5256.2023.06.029. [5] WEN X, KLIONSKY DJ. At a glance: A history of autophagy and cancer[J]. Semin Cancer Biol, 2020, 66: 3- 11. DOI: 10.1016/j.semcancer.2019.11.005. [6] FILALI-MOUNCEF Y, HUNTER C, ROCCIO F, et al. The ménage à trois of autophagy, lipid droplets and liver disease[J]. Autophagy, 2022, 18( 1): 50- 72. DOI: 10.1080/15548627.2021.1895658. [7] KOCAK M, EZAZI ERDI S, JORBA G, et al. Targeting autophagy in disease: established and new strategies[J]. Autophagy, 2022, 18( 3): 473- 495. DOI: 10.1080/15548627.2021.1936359. [8] YUAN C, LIAN QH, NI BB, et al. Screening and bioinformatics analysis of key autophagy-related genes in alcoholic hepatitis[J]. Ogran Transplant, 2024, 15( 1): 90- 101. DOI: 10.3969/j.issn.1674-7445.2023163.袁超, 练庆海, 尼贝贝, 等. 酒精性肝炎自噬关键基因的筛选及生物信息学分析[J]. 器官移植, 2024, 15( 1): 90- 101. DOI: 10.3969/j.issn.1674-7445.2023163. [9] KIRCHNER P, BOURDENX M, MADRIGAL-MATUTE J, et al. Proteome-wide analysis of chaperone-mediated autophagy targeting motifs[J]. PLoS Biol, 2019, 17( 5): e3000301. DOI: 10.1371/journal.pbio.3000301. [10] SAHU R, KAUSHIK S, CLEMENT CC, et al. Microautophagy of cytosolic proteins by late endosomes[J]. Dev Cell, 2011, 20( 1): 131- 139. DOI: 10.1016/j.devcel.2010.12.003. [11] WANG L, KLIONSKY DJ, SHEN HM. The emerging mechanisms and functions of microautophagy[J]. Nat Rev Mol Cell Biol, 2023, 24( 3): 186- 203. DOI: 10.1038/s41580-022-00529-z. [12] OLZMANN JA, CARVALHO P. Dynamics and functions of lipid droplets[J]. Nat Rev Mol Cell Biol, 2019, 20( 3): 137- 155. DOI: 10.1038/s41580-018-0085-z. [13] CHUNG J, PARK J, LAI ZW, et al. The Troyer syndrome protein spartin mediates selective autophagy of lipid droplets[J]. Nat Cell Biol, 2023, 25( 8): 1101- 1110. DOI: 10.1038/s41556-023-01178-w. [14] BACKE SJ, SAGER RA, HERITZ JA, et al. Activation of autophagy depends on Atg1/Ulk1-mediated phosphorylation and inhibition of the Hsp90 chaperone machinery[J]. Cell Rep, 2023, 42( 7): 112807. DOI: 10.1016/j.celrep.2023.112807. [15] XU Y, WANG S, LEUNG CK, et al. α-amanitin induces autophagy through AMPK-mTOR-ULK1 signaling pathway in hepatocytes[J]. Toxicol Lett, 2023, 383: 89- 97. DOI: 10.1016/j.toxlet.2023.06.004. [16] BAI J, ZHU Y, HE L, et al. Saponins from bitter melon reduce lipid accumulation via induction of autophagy in C. elegans and HepG2 cell line[J]. Curr Res Food Sci, 2022, 5: 1167- 1175. DOI: 10.1016/j.crfs.2022.06.011. [17] NGUYEN LN, BORMANN J, LE GT, et al. Autophagy-related lipase FgATG15 of Fusarium graminearum is important for lipid turnover and plant infection[J]. Fungal Genet Biol, 2011, 48( 3): 217- 224. DOI: 10.1016/j.fgb.2010.11.004. [18] KURUSU T, KOYANO T, HANAMATA S, et al. OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development[J]. Autophagy, 2014, 10( 5): 878- 888. DOI: 10.4161/auto.28279. [19] de la BALLINA LR, MUNSON MJ, SIMONSEN A. Lipids and lipid-binding proteins in selective autophagy[J]. J Mol Biol, 2020, 432( 1): 135- 159. DOI: 10.1016/j.jmb.2019.05.051. [20] BARROS J, MAGEN S, LAPIDOT-COHEN T, et al. Autophagy is required for lipid homeostasis during dark-induced senescence[J]. Plant Physiol, 2021, 185( 4): 1542- 1558. DOI: 10.1093/plphys/kiaa120. [21] MALLÉN-PONCE MJ, GÁMEZ-ARCAS S, PÉREZ-PÉREZ ME. Redox partner interactions in the ATG8 lipidation system in microalgae[J]. Free Radic Biol Med, 2023, 203: 58- 68. DOI: 10.1016/j.freeradbiomed.2023.04.004. [22] OUYANG Q, LIU R. MTOR-mediates hepatic lipid metabolism through an autophagic SNARE complex[J]. Autophagy, 2022, 18( 6): 1467- 1469. DOI: 10.1080/15548627.2022.2037853. [23] ADNAN M, ISLAM W, ZHANG J, et al. Diverse role of SNARE protein Sec22 in vesicle trafficking, membrane fusion, and autophagy[J]. Cells, 2019, 8( 4): 337. DOI: 10.3390/cells8040337. [24] SHROFF A, NAZARKO TY. SQSTM1, lipid droplets and current state of their lipophagy affairs[J]. Autophagy, 2023, 19( 2): 720- 723. DOI: 10.1080/15548627.2022.2094606. [25] SCHULZE RJ, DRIŽYTĖ K, CASEY CA, et al. Hepatic lipophagy: new insights into autophagic catabolism of lipid droplets in the liver[J]. Hepatol Commun, 2017, 1( 5): 359- 369. DOI: 10.1002/hep4.1056. [26] YANG M, LUO S, CHEN W, et al. Chaperone-mediated autophagy: a potential target for metabolic diseases[J]. Curr Med Chem, 2023, 30( 16): 1887- 1899. DOI: 10.2174/0929867329666220811141955. [27] SEO AY, LAU PW, FELICIANO D, et al. AMPK and vacuole-associated Atg14p orchestrate μ-lipophagy for energy production and long-term survival under glucose starvation[J]. Elife, 2017, 6: e21690. DOI: 10.7554/eLife.21690. [28] OKU M, MAEDA Y, KAGOHASHI Y, et al. Evidence for ESCRT- and clathrin-dependent microautophagy[J]. J Cell Biol, 2017, 216( 10): 3263- 3274. DOI: 10.1083/jcb.201611029. [29] HOMMA Y, HIRAGI S, FUKUDA M. Rab family of small GTPases: an updated view on their regulation and functions[J]. FEBS J, 2021, 288( 1): 36- 55. DOI: 10.1111/febs.15453. [30] LI Z, WELLER SG, DRIZYTE-MILLER K, et al. Maturation of lipophagic organelles in hepatocytes is dependent upon a Rab10/dynamin-2 complex[J]. Hepatology, 2020, 72( 2): 486- 502. DOI: 10.1002/hep.31059. [31] DENG Y, ZHOU C, MIRZA AH, et al. Rab18 binds PLIN2 and ACSL3 to mediate lipid droplet dynamics[J]. Biochim Biophys Acta Mol Cell Biol Lipids, 2021, 1866( 7): 158923. DOI: 10.1016/j.bbalip.2021.158923. [32] KLOSKA A, WĘSIERSKA M, MALINOWSKA M, et al. Lipophagy and lipolysis status in lipid storage and lipid metabolism diseases[J]. Int J Mol Sci, 2020, 21( 17). DOI: 10.3390/ijms21176113. [33] SINGH R, KAUSHIK S, WANG Y, et al. Autophagy regulates lipid metabolism[J]. Nature, 2009, 458( 7242): 1131- 1135. DOI: 10.1038/nature07976. [34] TAN YM, TAN YF, MENG GZ, et al. The regulatory role of lipophagy in lipid metabolism diseases[J]. J Med Sci Cent South China, 2022, 50( 5): 777- 780. DOI: 10.15972/j.cnki.43-1509/r.2022.05.039.谭艳美, 谭艳飞, 蒙国照, 等. 脂噬在脂代谢疾病中的调控作用[J]. 中南医学科学杂志, 2022, 50( 5): 777- 780. DOI: 10.15972/j.cnki.43-1509/r.2022.05.039. [35] CUI W, SATHYANARAYAN A, LOPRESTI M, et al. Lipophagy-derived fatty acids undergo extracellular efflux via lysosomal exocytosis[J]. Autophagy, 2021, 17( 3): 690- 705. DOI: 10.1080/15548627.2020.1728097. [36] ZHAO N, TAN H, WANG L, et al. Palmitate induces fat accumulation via repressing FoxO1-mediated ATGL-dependent lipolysis in HepG2 hepatocytes[J]. PLoS One, 2021, 16( 1): e0243938. DOI: 10.1371/journal.pone.0243938. [37] SATHYANARAYAN A, MASHEK MT, MASHEK DG. ATGL promotes autophagy/lipophagy via SIRT1 to control hepatic lipid droplet catabolism[J]. Cell Rep, 2017, 19( 1): 1- 9. DOI: 10.1016/j.celrep.2017.03.026. [38] ZHANG G, HAN J, WANG L, et al. The vesicular transporter STX11 governs ATGL-mediated hepatic lipolysis and lipophagy[J]. iScience, 2022, 25( 4): 104085. DOI: 10.1016/j.isci.2022.104085. [39] LI L, LI Q, HUANG W, et al. Dapagliflozin alleviates hepatic steatosis by restoring autophagy via the AMPK-mTOR pathway[J]. Front Pharmacol, 2021, 12: 589273. DOI: 10.3389/fphar.2021.589273. [40] ZHANG D, ZHANG Y, WANG Z, et al. Thymoquinone attenuates hepatic lipid accumulation by inducing autophagy via AMPK/mTOR/ULK1-dependent pathway in nonalcoholic fatty liver disease[J]. Phytother Res, 2023, 37( 3): 781- 797. DOI: 10.1002/ptr.7662. [41] SEOK S, KIM YC, BYUN S, et al. Fasting-induced JMJD3 histone demethylase epigenetically activates mitochondrial fatty acid β-oxidation[J]. J Clin Invest, 2018, 128( 7): 3144- 3159. DOI: 10.1172/JCI97736. [42] TALUKDAR S, KHARITONENKOV A. FGF19 and FGF21: In NASH we trust[J]. Mol Metab, 2021, 46: 101152. DOI: 10.1016/j.molmet.2020.101152. [43] BYUN S, SEOK S, KIM YC, et al. Fasting-induced FGF21 signaling activates hepatic autophagy and lipid degradation via JMJD3 histone demethylase[J]. Nat Commun, 2020, 11( 1): 807. DOI: 10.1038/s41467-020-14384-z. [44] ALSHEHADE S, ALSHAWSH MA, MURUGAIYAH V, et al. The role of protein kinases as key drivers of metabolic dysfunction-associated fatty liver disease progression: New insights and future directions[J]. Life Sci, 2022, 305: 120732. DOI: 10.1016/j.lfs.2022.120732. [45] LIU YY, SUI M, JIANG XF, et al. Effect of Danzhi Tiaozhi decoction on the PI3K/AKT/FOXO1 signaling pathway in high-fat induced MAFLD rats[J]. J Nangjing Univ Tradit Chin Med, 2023, 39( 6): 541- 547. DOI: 10.14148/j.issn.1672-0482.2023.0541.刘玉玉, 隋淼, 蒋小飞, 等. 丹栀调脂汤对高脂诱导MAFLD大鼠PI3K/AKT/FOXO1信号通路的影响[J]. 南京中医药大学学报, 2023, 39( 6): 541- 547. DOI: 10.14148/j.issn.1672-0482.2023.0541. [46] WANG S, YANG FJ, SHANG LC, et al. Puerarin protects against high-fat high-sucrose diet-induced non-alcoholic fatty liver disease by modulating PARP-1/PI3K/AKT signaling pathway and facilitating mitochondrial homeostasis[J]. Phytother Res, 2019, 33( 9): 2347- 2359. DOI: 10.1002/ptr.6417. [47] WANG MY, LI EW, GAO G, et al. Zexie Decoction regulates Akt/TFEB signaling pathway to promote lipophagy in hepatocytes[J]. China J Chin Mater Med, 2022, 47( 22): 6183- 6190. DOI: 10.19540/j.cnki.cjcmm.20220706.702.王梦瑶, 李二稳, 高改, 等. 泽泻汤调控Akt/TFEB促进肝细胞脂噬机制研究[J]. 中国中药杂志, 2022, 47( 22): 6183- 6190. DOI: 10.19540/j.cnki.cjcmm.20220706.702. [48] YAN H, CHAI CY, ZHANG D, et al. Explore the mechanism of autophagy and insulin resistance in non-alcoholic fatty liver disease based on JNK signaling pathway[J]. Shaanxi Med J, 2023, 52( 11): 1506- 1510. DOI: 10.3969/j.issn.1000-7377.2023.11.012.延华, 柴春艳, 张丹, 等. 基于JNK信号通路探讨自噬、胰岛素抵抗在非酒精性脂肪性肝病中的发病机制[J]. 陕西医学杂志, 2023, 52( 11): 1506- 1510. DOI: 10.3969/j.issn.1000-7377.2023.11.012. [49] GONG J, GAO X, GE S, et al. The role of cGAS-STING signalling in metabolic diseases: from signalling networks to targeted intervention[J]. Int J Biol Sci, 2024, 20( 1): 152- 174. DOI: 10.7150/ijbs.84890. [50] PANZITT K, WAGNER M. FXR in liver physiology: Multiple faces to regulate liver metabolism[J]. Biochim Biophys Acta Mol Basis Dis, 2021, 1867( 7): 166133. DOI: 10.1016/j.bbadis.2021.166133. [51] MA SY, SUN KS, ZHANG M, et al. Disruption of Plin5 degradation by CMA causes lipid homeostasis imbalance in NAFLD[J]. Liver Int, 2020, 40( 10): 2427- 2438. DOI: 10.1111/liv.14492. [52] YOU Y, LI WZ, ZHANG S, et al. SNX10 mediates alcohol-induced liver injury and steatosis by regulating the activation of chaperone-mediated autophagy[J]. J Hepatol, 2018, 69( 1): 129- 141. DOI: 10.1016/j.jhep.2018.01.038. [53] LEE W, KIM HY, CHOI YJ, et al. SNX10-mediated degradation of LAMP2A by NSAIDs inhibits chaperone-mediated autophagy and induces hepatic lipid accumulation[J]. Theranostics, 2022, 12( 5): 2351- 2369. DOI: 10.7150/thno.70692. [54] QIAO L, HU J, QIU X, et al. LAMP2A, LAMP2B and LAMP2C: similar structures, divergent roles[J]. Autophagy, 2023, 19( 11): 2837- 2852. DOI: 10.1080/15548627.2023.2235196. [55] SCHULZE RJ, KRUEGER EW, WELLER SG, et al. Direct lysosome-based autophagy of lipid droplets in hepatocytes[J]. Proc Natl Acad Sci U S A, 2020, 117( 51): 32443- 32452. DOI: 10.1073/pnas.2011442117. [56] LIAO PC, GARCIA EJ, TAN G, et al. Roles for Lo microdomains and ESCRT in ER stress-induced lipid droplet microautophagy in budding yeast[J]. Mol Biol Cell, 2021, 32( 22): br12. DOI: 10.1091/mbc.E21-04-0179. [57] GARCIA EJ, LIAO PC, TAN G, et al. Membrane dynamics and protein targets of lipid droplet microautophagy during ER stress-induced proteostasis in the budding yeast, Saccharomyces cerevisiae[J]. Autophagy, 2021, 17( 9): 2363- 2383. DOI: 10.1080/15548627.2020.1826691. -



 PDF下载 ( 1212 KB)
PDF下载 ( 1212 KB)

 下载:
下载: