免疫原性细胞死亡基因风险模型的建立及其对肝细胞癌预后和肿瘤微环境特征的预测价值
DOI: 10.12449/JCH241218
Establishment of a risk model based on immunogenic cell death-related genes and its value in predicting the prognosis and tumor microenvironment characteristics of hepatocellular carcinoma
-
摘要:
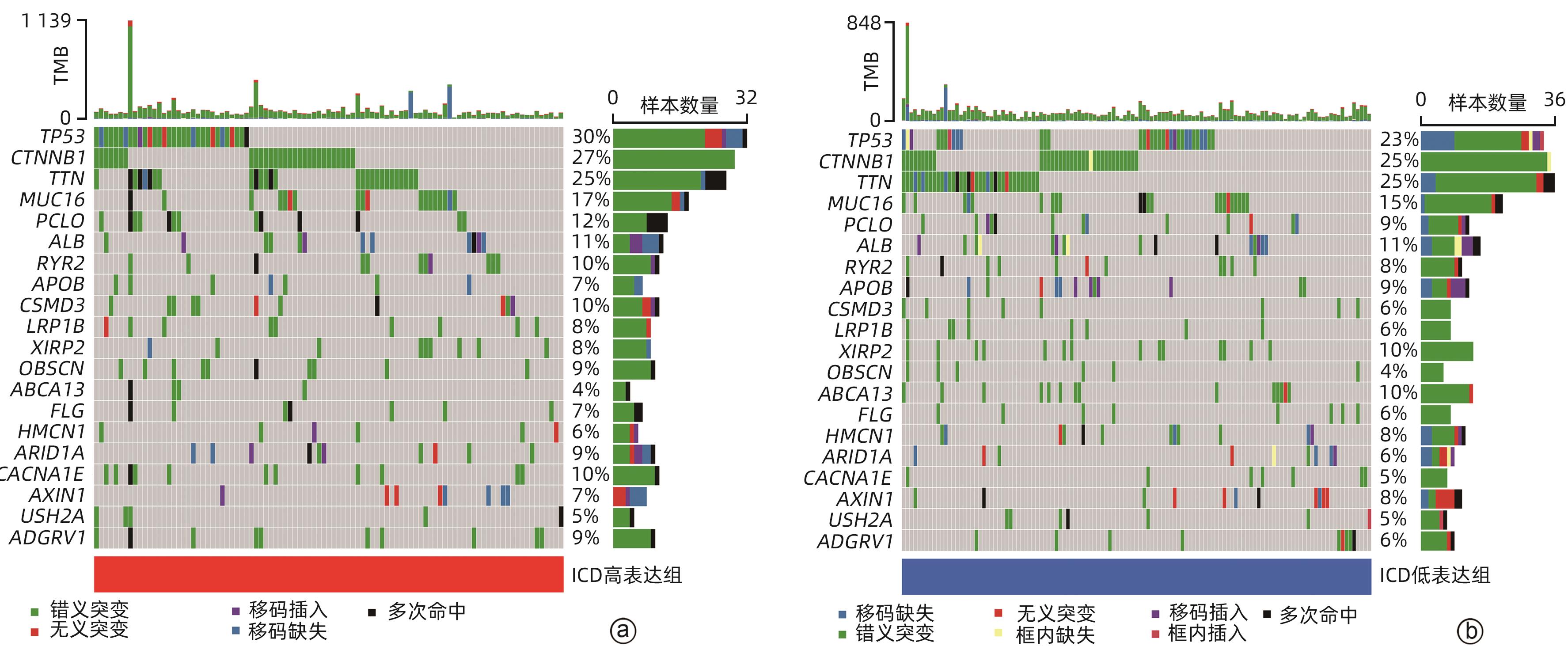
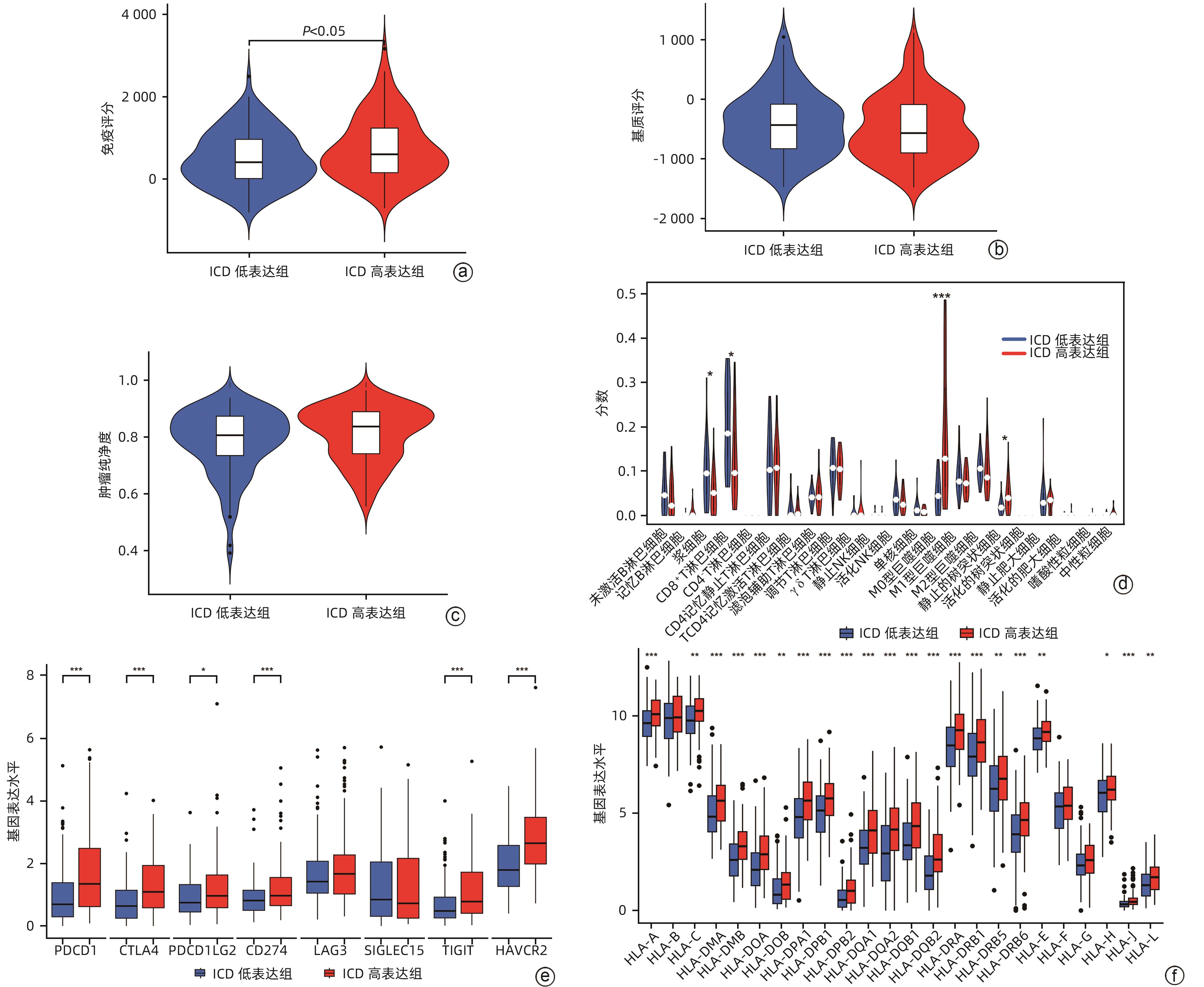
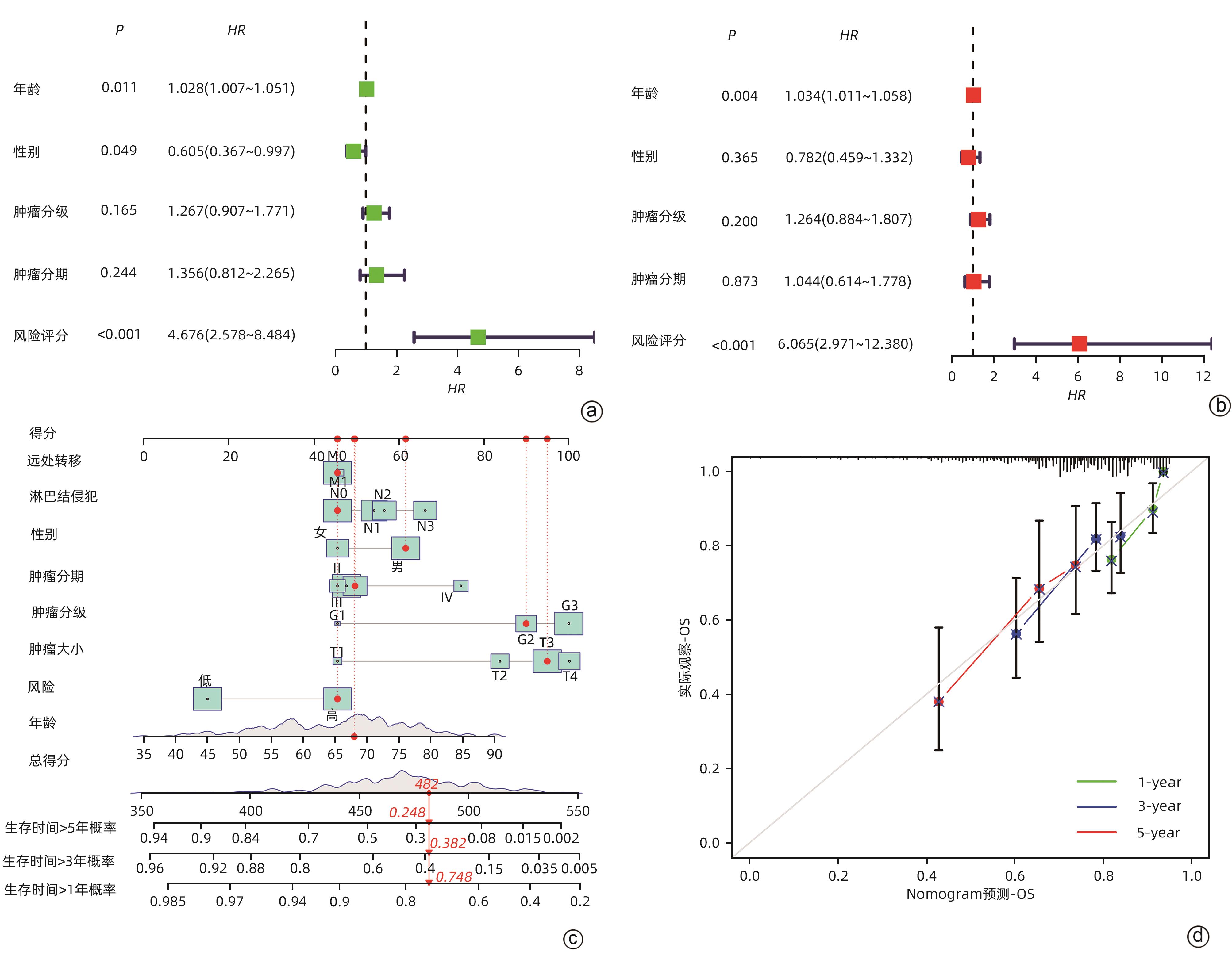
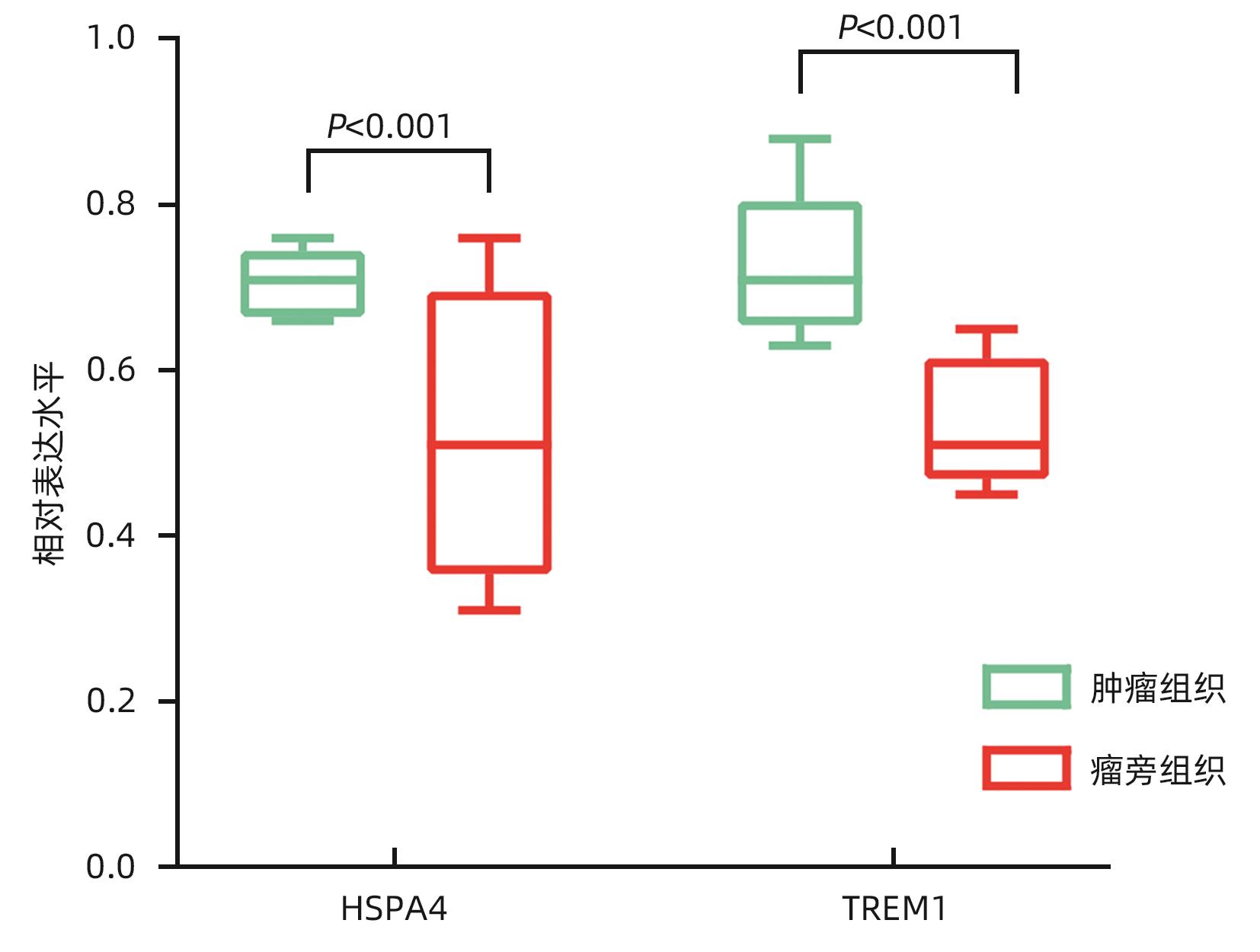
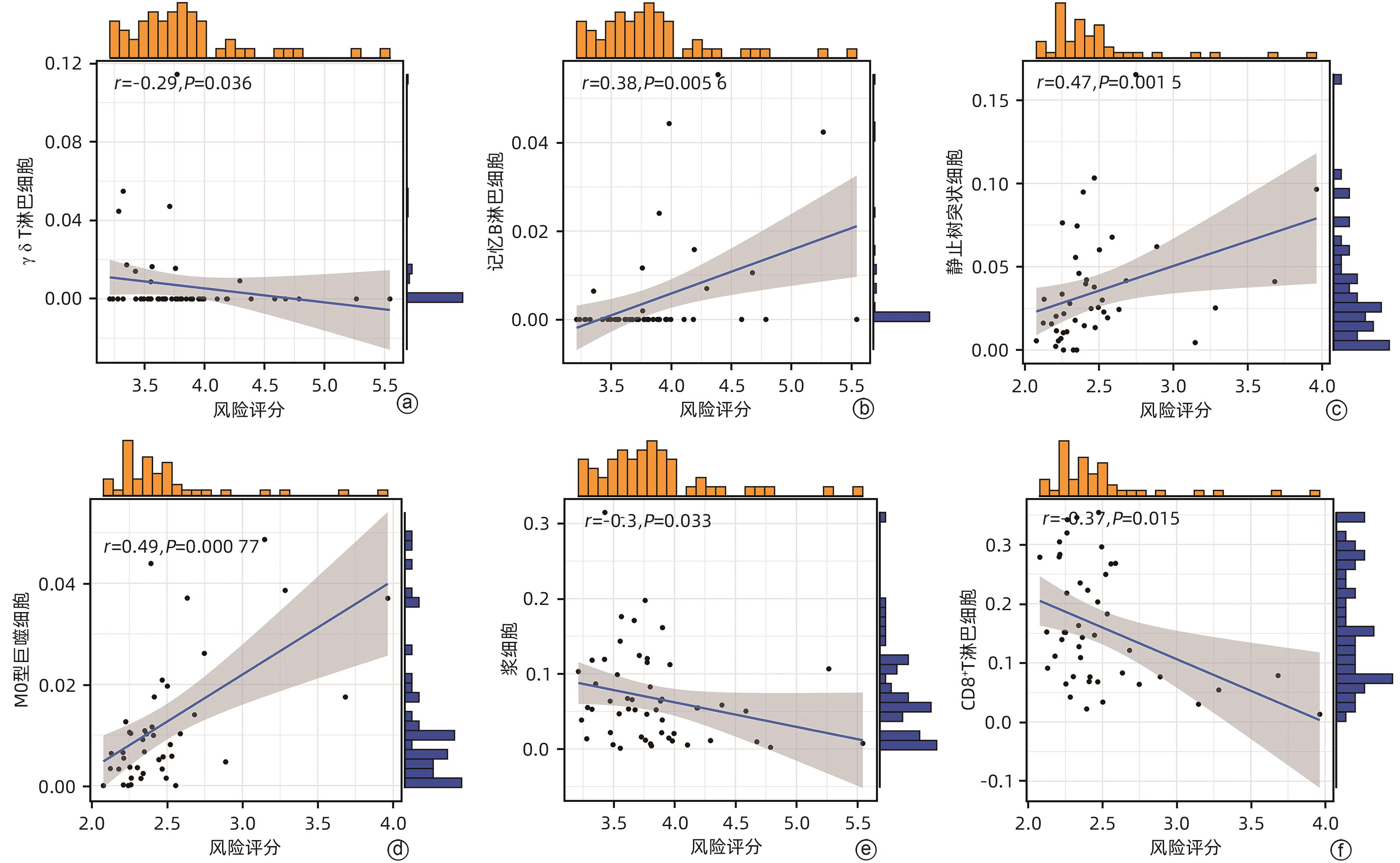
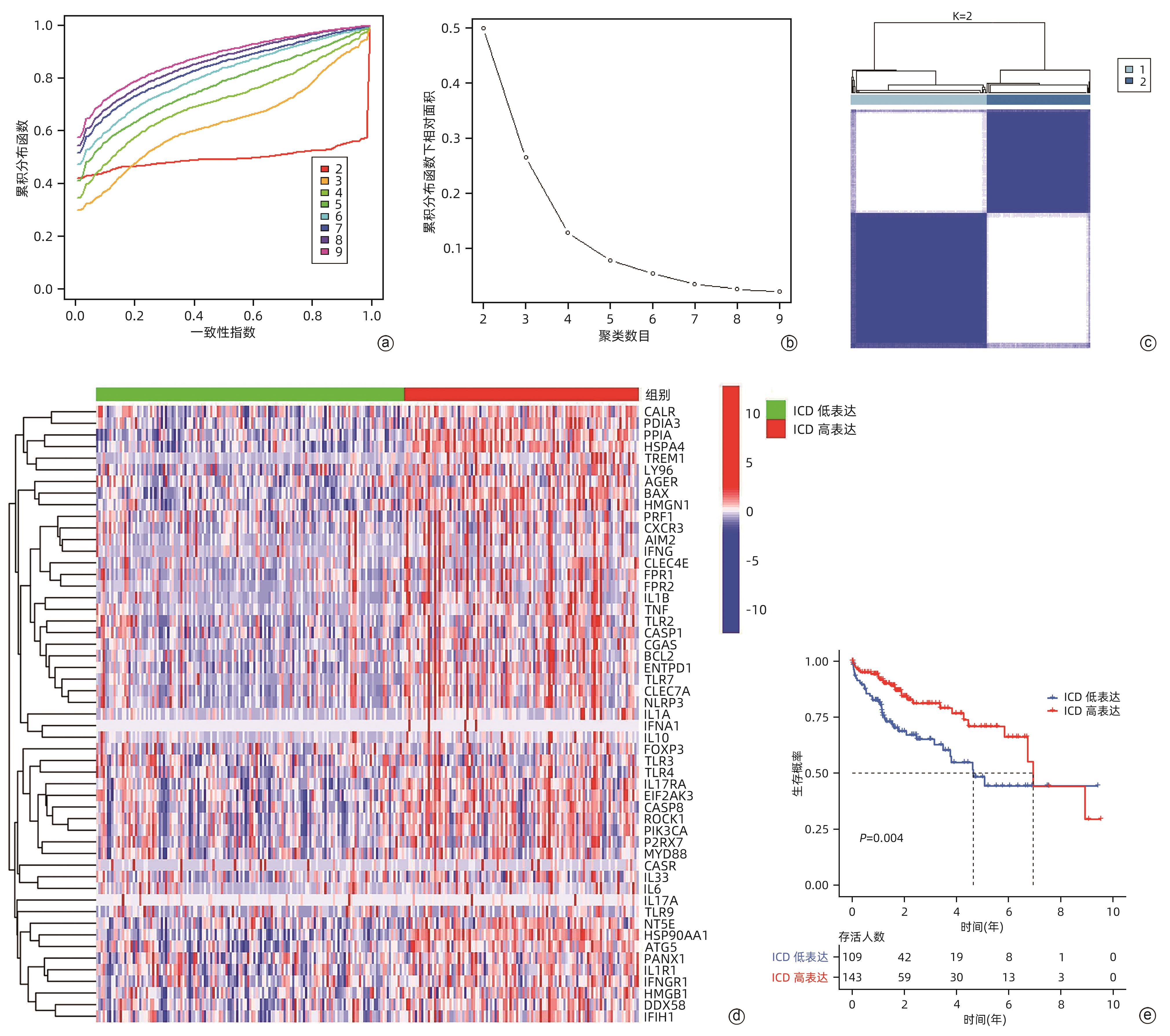
目的 鉴定肝细胞癌(HCC)免疫原性细胞死亡(ICD)相关基因,并基于相关基因构建评分模型预测HCC的预后和肿瘤微环境特征。 方法 通过癌症基因组图谱数据库获取HCC数据集,采用热图展示了HCC中57个ICD相关基因的表达。基于ICD相关基因表达进行聚类分析,对2种ICD相关亚型(ICD低表达组和ICD高表达组)进行基因本体富集、京都基因和基因组百科全书富集、体细胞突变差异、免疫细胞浸润差异分析。构建LASSO Cox回归风险预后模型,验证其临床应用价值。构建列线图模型,预测患者1、3和5年的生存率。此外,采用qRT-PCR验证模型中关键基因的表达水平。两组间比较采用成组t检验,采用单因素和多因素Cox回归分析确定临床病理特征中的预后因素。采用Kaplan-Meier生存曲线进行预后分析。采用Spearman秩相关进行相关性分析。 结果 ICD低表达组预后较差,ICD高表达组与良好的临床结果相关(P=0.004)。进一步研究表明ICD高表达组与免疫活性微环境相关,且ICD高表达组的基因主要富集于免疫相关通路(免疫球蛋白受体结合、造血细胞谱系和B淋巴细胞受体信号通路)。体细胞突变结果显示,ICD高表达组的CD274、CTLA4、HAVCR2、TIGIT、PDCD1和PDCD1LG2的基因表达水平较高(P值均<0.05)。利用8个ICD相关基因(HSP90AA1、ATG5、BAX、PPIA、HSPA4、TLR2、TREM1、LY96)建立风险预后模型,该模型在不同临床特征中均有较好的预测价值。单因素和多因素Cox回归分析表明,在训练集中,年龄和风险评分是总生存期的独立预后因素(P值均<0.05)。qRT-PCR结果表明,HSPA4和REM1在HCC肿瘤样本中的相对表达水平显著高于瘤旁组织(P值均<0.001)。ICD风险评分升高的患者与γδT淋巴细胞(r=-0.29)、浆细胞(r=-0.3)和CD8+T淋巴细胞(r=-0.37)呈负相关(P值均<0.05),与记忆B淋巴细胞(r=0.38)、静止树突状细胞(r=0.47)和M0型巨噬细胞(r=0.49)呈正相关(P值均<0.05)。 结论 本研究确定了与HCC预后相关的ICD基因,为理解不同ICD表达谱相关的免疫特性提供了见解。构建的风险模型和列线图对于预测HCC患者的预后预测和免疫治疗指导具有重要的价值。 Abstract:Objective To identify immunogenic cell death (ICD)-related genes in hepatocellular carcinoma (HCC), and to establish a scoring model based on these genes for predicting the prognosis and tumor microenvironment characteristics of HCC. Methods The Cancer Genome Atlas database was used to obtain HCC datasets, and heatmaps were used to display the expression of 57 ICD-related genes in HCC. A cluster analysis was conducted based on the expression of ICD-related genes, and two ICD subtypes (low and high ICD expression groups) were analyzed in terms of gene ontology enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis, somatic mutation, and immune cell infiltration. The LASSO Cox regression risk model was constructed to evaluate its clinical application value, and a nomogram model was established to predict the 1-, 3-, and 5-year survival rates of patients. In addition, qRT-PCR was used to validate the expression levels of key genes in the model. The independent-samples t test was used for comparison between two groups, and the univariate and multivariate Cox regression analyses were used to determine prognostic factors among clinicopathological features. The Kaplan-Meier survival curve was used for prognostic analysis, and the Spearman rank correlation test was used for correlation analysis. Results The low ICD expression group had a poorer prognosis, while the high ICD expression group had relatively favorable clinical outcomes (P=0.004). Further analysis showed that the high ICD expression group was associated with an immune-active microenvironment, and the genes were mainly enriched in immune-related pathways such as immunoglobulin receptor binding, hematopoietic cell lineage, and B cell receptor. The results of somatic mutation analysis showed that the high ICD expression group had higher expression levels of CD274, CTLA4, HAVCR2, TIGIT, PDCD1, and PDCD1LG2 (all P<0.05). A risk prediction model was established using 8 ICD-related genes, i.e., HSP90AA1, ATG5, BAX, PPIA, HSPA4, TLR2, TREM1, and LY96, and this model showed a good predictive value across different clinical characteristics. The univariate and multivariate Cox regression analyses showed that age and risk score were independent prognostic factors for overall survival in the training set (both P<0.05). The results of qRT-PCR showed that the relative expression levels of HSPA4 and REM1 in HCC tumor samples were significantly higher than those in adjacent tissue samples (both P<0.001). For the patients with an increase in ICD risk score, the ICD risk score was negatively correlated with γδT cells (r=-0.29, P<0.05), plasma cells (r=-0.3, P<0.05), and CD8+T lymphocytes (r=-0.37, P<0.05) and was positively correlated with memory B cells (r=0.38, P<0.05), resting dendritic cells (r=0.47, P<0.05), and M0 macrophages (r=0.49, P<0.05). Conclusion This study identifies the ICD-related genes that are associated with the prognosis of HCC, which provides insights into the immune characteristics of different ICD expression profiles. The risk model and the nomogram model established in this study have a significant value for predicting the prognosis of HCC patients and guiding immunotherapy for HCC patients. -
Key words:
- Carcinoma, Hepatocellular /
- Immunogenic Cell Death /
- Prognosis /
- Tumor Microenvironment
-
表 1 57个ICD相关基因
Table 1. 57 ICD-related genes
基因 ENTPD1、NT5E、CALR、HMGB1、HSP90AA1、ATG5、BAX、CASP8、CASR、TLR3、TLR7、TLR9、CLEC4E、CLEC7A、DDX58、IFIH1、CGAS、IL1A、IL33、ROCK1、PANX1、BCL2、PPIA、HSPA4、TLR2、AIM2、AGER、TREM1、FPR1、FPR2、P2Y2R、P2Y6R、P2Y12R、IFNA1、CASP1、IL1R1、IL1B、NLRP3、P2RX7、LY96、MYD88、PDIA3、EIF2AK3、PIK3CA、FNB1、CXCR3、IL10、IL6、TNF、TLR4、FOXP3、IFNG、IFNGR1、IL17A、IL17RA、PRF1、HMGN1 表 2 HSPA4和TREM1基因的正、反向引物
Table 2. Forward and reverse primers for HSPA4 and TREM1 genes
名称 序列 HSPA4 正向引物:5'-ACCAGGACACTGTTGAGTTC-3' 反向引物:5'-ACTCATCTCCGAGTTCACAC-3' TREM1 正向引物:5'-TGCGGTTGTTTCCTCTCCTGGTCTTG-3' 反向引物:5'-TGTGAAATAGACACCGCTGAAGGTC‑ACT-3' -
[1] VOGEL A, MEYER T, SAPISOCHIN G, et al. Hepatocellular carcinoma[J]. Lancet, 2022, 400: 1345- 1362. DOI: 10.1016/S0140-6736(22)01200-4. [2] GANESAN P, KULIK LM. Hepatocellular carcinoma: New developments[J]. Clin Liver Dis, 2023, 27( 1): 85- 102. DOI: 10.1016/j.cld.2022.08.004. [3] LLOVET JM, KELLEY RK, VILLANUEVA A, et al. Hepatocellular carcinoma[J]. Nat Rev Dis Primers, 2021, 7( 1): 6. DOI: 10.1038/s41572-020-00240-3. [4] ZHANG YY, ZHANG ZM. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications[J]. Cell Mol Immunol, 2020, 17( 8): 807- 821. DOI: 10.1038/s41423-020-0488-6. [5] NAIMI A, MOHAMMED RN, RAJI A, et al. Tumor immunotherapies by immune checkpoint inhibitors(ICIs); the pros and cons[J]. Cell Commun Signal, 2022, 20( 1): 44. DOI: 10.1186/s12964-022-00854-y. [6] KENNEDY LB, SALAMA AKS. A review of cancer immunotherapy toxicity[J]. CA Cancer J Clin, 2020, 70( 2): 86- 104. DOI: 10.3322/caac.21596. [7] LLOVET JM, CASTET F, HEIKENWALDER M, et al. Immunotherapies for hepatocellular carcinoma[J]. Nat Rev Clin Oncol, 2022, 19( 3): 151- 172. DOI: 10.1038/s41571-021-00573-2. [8] SANGRO B, SAROBE P, HERVÁS-STUBBS S, et al. Advances in immunotherapy for hepatocellular carcinoma[J]. Nat Rev Gastroenterol Hepatol, 2021, 18( 8): 525- 543. DOI: 10.1038/s41575-021-00438-0. [9] NIU X, CHEN LJ, LI Y, et al. Ferroptosis, necroptosis, and pyroptosis in the tumor microenvironment: Perspectives for immunotherapy of SCLC[J]. Semin Cancer Biol, 2022, 86( Pt 3): 273- 285. DOI: 10.1016/j.semcancer.2022.03.009. [10] CAI JY, HU YY, YE Z, et al. Immunogenic cell death-related risk signature predicts prognosis and characterizes the tumour microenvironment in lower-grade glioma[J]. Front Immunol, 2022, 13: 1011757. DOI: 10.3389/fimmu.2022.1011757. [11] KROEMER G, GALASSI C, ZITVOGEL L, et al. Immunogenic cell stress and death[J]. Nat Immunol, 2022, 23( 4): 487- 500. DOI: 10.1038/s41590-022-01132-2. [12] AHMED A, TAIT SWG. Targeting immunogenic cell death in cancer[J]. Mol Oncol, 2020, 14( 12): 2994- 3006. DOI: 10.1002/1878-0261.12851. [13] FUCIKOVA J, KEPP O, KASIKOVA L, et al. Detection of immunogenic cell death and its relevance for cancer therapy[J]. Cell Death Dis, 2020, 11( 11): 1013. DOI: 10.1038/s41419-020-03221-2. [14] ZHOU JY, WANG GY, CHEN YZ, et al. Immunogenic cell death in cancer therapy: Present and emerging inducers[J]. J Cell Mol Med, 2019, 23( 8): 4854- 4865. DOI: 10.1111/jcmm.14356. [15] CHIARAVALLI M, SPRING A, AGOSTINI A, et al. Immunogenic cell death: An emerging target in gastrointestinal cancers[J]. Cells, 2022, 11( 19): 3033. DOI: 10.3390/cells11193033. [16] GALLUZZI L, KEPP O, HETT E, et al. Immunogenic cell death in cancer: Concept and therapeutic implications[J]. J Transl Med, 2023, 21( 1): 162. DOI: 10.1186/s12967-023-04017-6. [17] WANG XW, WU SW, LIU F, et al. An immunogenic cell death-related classification predicts prognosis and response to immunotherapy in head and neck squamous cell carcinoma[J]. Front Immunol, 2021, 12: 781466. DOI: 10.3389/fimmu.2021.781466. [18] GARG AD, DE RUYSSCHER D, AGOSTINIS P. Immunological metagene signatures derived from immunogenic cancer cell death associate with improved survival of patients with lung, breast or ovarian malignancies: A large-scale meta-analysis[J]. Oncoimmunology, 2015, 5( 2): e1069938. DOI: 10.1080/2162402X.2015.1069938. [19] BROWN ZJ, TSILIMIGRAS DI, RUFF SM, et al. Management of hepatocellular carcinoma: A review[J]. JAMA Surg, 2023, 158( 4): 410- 420. DOI: 10.1001/jamasurg.2022.7989. [20] TRICOLI L, NITURE S, CHIMEH U, et al. Role of microRNAs in the development of hepatocellular carcinoma and drug resistance[J]. Front Biosci(Landmark Ed), 2019, 24( 2): 382- 391. DOI: 10.2741/4724. [21] CEBALLOS MP, QUIROGA AD, PALMA NF. Role of sirtuins in hepatocellular carcinoma progression and multidrug resistance: Mechanistical and pharmacological perspectives[J]. Biochem Pharmacol, 2023, 212: 115573. DOI: 10.1016/j.bcp.2023.115573. [22] DEVAN AR, KUMAR AR, NAIR B, et al. Insights into an immunotherapeutic approach to combat multidrug resistance in hepatocellular carcinoma[J]. Pharmaceuticals(Basel), 2021, 14( 7): 656. DOI: 10.3390/ph14070656. [23] LI ZL, LAI XQ, FU SQ, et al. Immunogenic cell death activates the tumor immune microenvironment to boost the immunotherapy efficiency[J]. Adv Sci(Weinh), 2022, 9( 22): e2201734. DOI: 10.1002/advs.202201734. [24] ZHANG SW, WANG J, KONG ZQ, et al. Emerging photodynamic nanotherapeutics for inducing immunogenic cell death and potentiating cancer immunotherapy[J]. Biomaterials, 2022, 282: 121433. DOI: 10.1016/j.biomaterials.2022.121433. [25] LIU GH, QIU YM, ZHANG P, et al. Immunogenic cell death enhances immunotherapy of diffuse intrinsic pontine glioma: From preclinical to clinical studies[J]. Pharmaceutics, 2022, 14( 9): 1762. DOI: 10.3390/pharmaceutics14091762. [26] SANSONE C, BRUNO A, PISCITELLI C, et al. Natural compounds of marine origin as inducers of immunogenic cell death(ICD): Potential role for cancer interception and therapy[J]. Cells, 2021, 10( 2): 231. DOI: 10.3390/cells10020231. -



 PDF下载 ( 6319 KB)
PDF下载 ( 6319 KB)

 下载:
下载: