氨基酸代谢重编程对肝细胞癌免疫微环境的影响
DOI: 10.12449/JCH241226
Effect of amino acid metabolic reprogramming on immune microenvironment of hepatocellular carcinoma
-
摘要: 肿瘤免疫微环境是由肿瘤免疫细胞及其分泌的细胞因子构成的肿瘤局部外环境,对肿瘤的发生和发展具有一定调控作用。在肝细胞癌的治疗中,氨基酸代谢及其对增殖细胞代谢的重编程日益受到关注,显示出在调控肿瘤免疫微环境中的潜力。尽管氨基酸代谢重编程被视为治疗肿瘤的新途径,但其在调控肝细胞癌中肿瘤免疫的具体机制尚未明确。本文深入探讨了氨基酸代谢在肝细胞癌肿瘤免疫微环境中的作用机制及其临床应用前景,旨在为肝癌的免疫治疗研究提供新的思路。Abstract: Tumor immune microenvironment is a local external tumor environment composed of tumor immune cells and the cytokines secreted by these cells, and it plays a regulatory role in the development and progression of tumors. In the treatment of hepatocellular carcinoma, amino acid metabolism and its reprogramming of proliferating cell metabolism have attracted more and more attention, showing potential in regulating the tumor immune microenvironment. Although amino acid metabolic reprogramming is regarded as a novel approach for tumor therapy, its specific mechanism remains unclear in the regulation of tumor immunity in hepatocellular carcinoma. This article discusses the mechanism of action of amino acid metabolism in the tumor immune microenvironment of hepatocellular carcinoma and its application prospect in clinical practice, in order to provide new ideas for immunotherapy for liver cancer.
-
Key words:
- Carcinoma, Hepatocellular /
- Tumor Microenvironment /
- Amino Acids /
- Metabolism /
- Immunomodulation
-
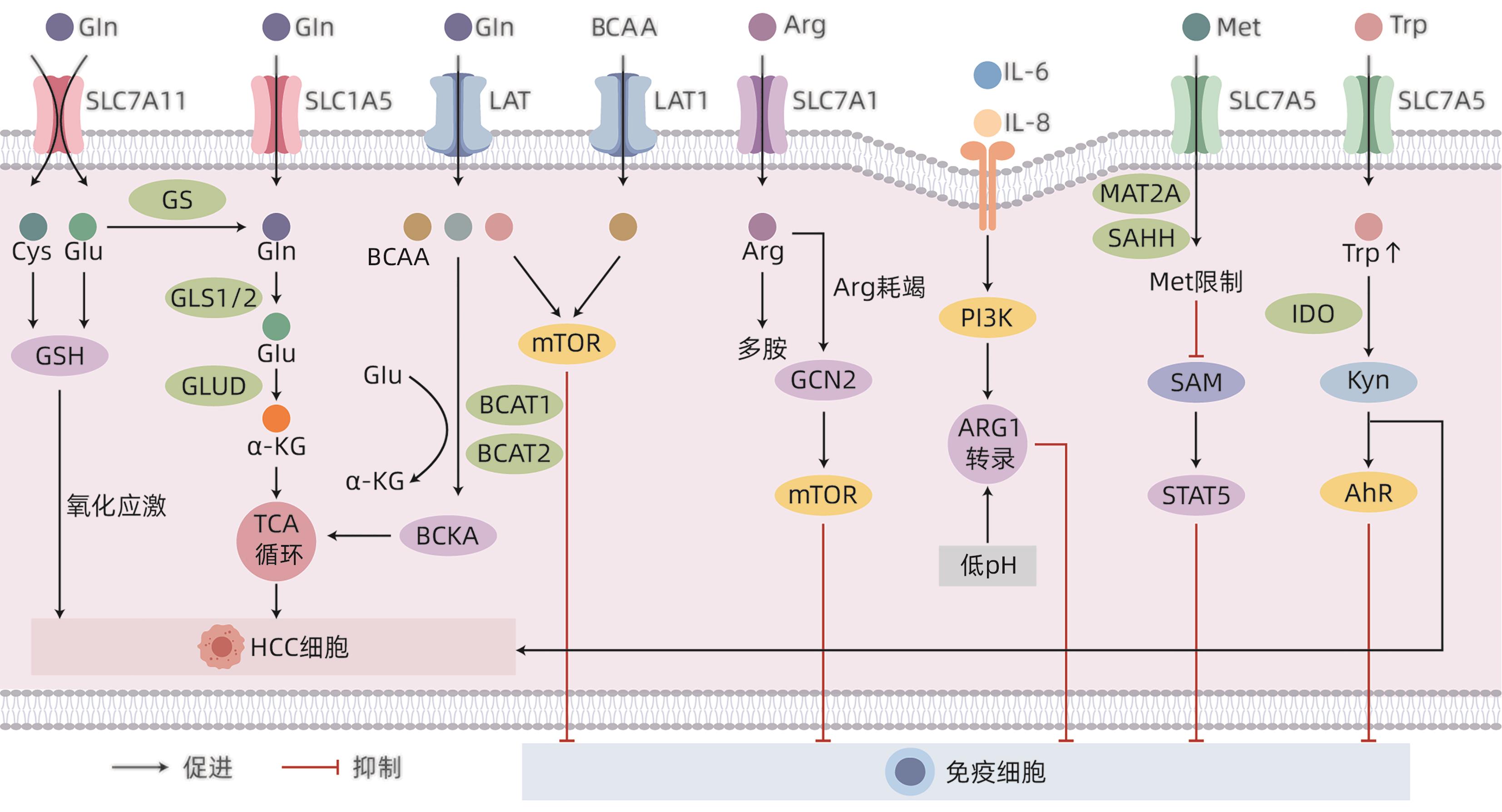
注: Cys,半胱氨酸;GLUD,谷氨酸脱氢酶;α-KG,α-酮戊二酸;BCAA,支链氨基酸;LAT,L型氨基酸转运蛋白;BCKA,支链氨基酸酮酸;Arg,精氨酸;PI3K,磷脂酰肌醇三激酶;MAT2A,Met腺苷转移酶Ⅱα;SAHH,S-腺苷同型半胱氨酸水解酶;SAM,S-腺苷甲硫氨酸;STAT5,信号传导及转录激活蛋白5。
图 1 氨基酸代谢重编程对HCC免疫微环境的影响
Figure 1. Effects of reprogramming amino acid metabolism on the immune microenvironment in hepatocellular carcinoma
-
[1] ZHENG YL, LI L, JIA YX, et al. LINC01554-mediated glucose metabolism reprogramming suppresses tumorigenicity in hepatocellular carcinoma via downregulating PKM2 expression and inhibiting Akt/mTOR signaling pathway[J]. Theranostics, 2019, 9( 3): 796- 810. DOI: 10.7150/thno.28992. [2] RUMGAY H, ARNOLD M, FERLAY J, et al. Global burden of primary liver cancer in 2020 and predictions to 2040[J]. J Hepatol, 2022, 77( 6): 1598- 1606. DOI: 10.1016/j.jhep.2022.08.021. [3] Department of Medical Emergency, National Health Commission of the People’s Republic of China. Healthy China action-implementation plan of cancer prevention action(2023-2030)[J]. China Cancer, 2023, 32( 12): 887- 890. DOI: 10.11735/j.issn.1004-0242.2023.12.A001.中华人民共和国国家卫生健康委员会医疗应急司. 健康中国行动——癌症防治行动实施方案(2023—2030年)[J]. 中国肿瘤, 2023, 32( 12): 887- 890. DOI: 10.11735/j.issn.1004-0242.2023.12.A001. [4] NONG SQ, HAN XY, XIANG Y, et al. Metabolic reprogramming in cancer: Mechanisms and therapeutics[J]. MedComm(2020), 2023, 4( 2): e218. DOI: 10.1002/mco2.218. [5] FOGLIA B, BELTRÀ M, SUTTI S, et al. Metabolic reprogramming of HCC: A new microenvironment for immune responses[J]. Int J Mol Sci, 2023, 24( 8): 7463. DOI: 10.3390/ijms24087463. [6] PAGET S. The distribution of secondary growths in cancer of the breast. 1889[J]. Cancer Metastasis Rev, 1989, 8( 2): 98- 101. [7] ZHANG Q, LOU Y, BAI XL, et al. Immunometabolism: A novel perspective of liver cancer microenvironment and its influence on tumor progression[J]. World J Gastroenterol, 2018, 24( 31): 3500- 3512. DOI: 10.3748/wjg.v24.i31.3500. [8] KOTSARI M, DIMOPOULOU V, KOSKINAS J, et al. Immune system and hepatocellular carcinoma(HCC): New insights into HCC progression[J]. Int J Mol Sci, 2023, 24( 14): 11471. DOI: 10.3390/ijms241411471. [9] PAULUSMA CC, LAMERS WH, BROER S, et al. Amino acid metabolism, transport and signalling in the liver revisited[J]. Biochem Pharmacol, 2022, 201: 115074. DOI: 10.1016/j.bcp.2022.115074. [10] ZHENG Y, YAO YR, GE TX, et al. Amino acid metabolism reprogramming: Shedding new light on T cell anti-tumor immunity[J]. J Exp Clin Cancer Res, 2023, 42( 1): 291. DOI: 10.1186/s13046-023-02845-4. [11] ZHANG XH, ZHANG SY, LI L. Amino acid metabolic reprogramming in tumorigenesis and development[J]. Chin J Biochem Mol Biol, 2023, 39( 2): 174- 188. DOI: 10.13865/j.cnki.cjbmb.2022.06.1105.张仙宏, 张思雨, 李乐. 氨基酸代谢重编程在肿瘤发生发展中的作用[J]. 中国生物化学与分子生物学报, 2023, 39( 2): 174- 188. DOI: 10.13865/j.cnki.cjbmb.2022.06.1105. [12] YANG F, HILAKIVI-CLARKE L, SHAHA A, et al. Metabolic reprogramming and its clinical implication for liver cancer[J]. Hepatology, 2023, 78( 5): 1602- 1624. DOI: 10.1097/HEP.0000000000000005. [13] WANG YY, BAI CS, RUAN YX, et al. Coordinative metabolism of glutamine carbon and nitrogen in proliferating cancer cells under hypoxia[J]. Nat Commun, 2019, 10( 1): 201. DOI: 10.1038/s41467-018-08033-9. [14] JIN HJ, WANG SY, ZAAL EA, et al. A powerful drug combination strategy targeting glutamine addiction for the treatment of human liver cancer[J]. eLife, 2020, 9: e56749. DOI: 10.7554/eLife.56749. [15] LIN J, RAO DN, ZHANG M, et al. Metabolic reprogramming in the tumor microenvironment of liver cancer[J]. J Hematol Oncol, 2024, 17( 1): 6. DOI: 10.1186/s13045-024-01527-8. [16] TIAN LY, SMIT DJ, JÜCKER M. The role of PI3K/AKT/mTOR signaling in hepatocellular carcinoma metabolism[J]. Int J Mol Sci, 2023, 24( 3): 2652. DOI: 10.3390/ijms24032652. [17] MERLIN J, IVANOV S, DUMONT A, et al. Non-canonical glutamine transamination sustains efferocytosis by coupling redox buffering to oxidative phosphorylation[J]. Nat Metab, 2021, 3( 10): 1313- 1326. DOI: 10.1038/s42255-021-00471-y. [18] MA GF, ZHANG ZL, LI P, et al. Reprogramming of glutamine metabolism and its impact on immune response in the tumor microenvironment[J]. Cell Commun Signal, 2022, 20( 1): 114. DOI: 10.1186/s12964-022-00909-0. [19] YE YY, YU BD, WANG H, et al. Glutamine metabolic reprogramming in hepatocellular carcinoma[J]. Front Mol Biosci, 2023, 10: 1242059. DOI: 10.3389/fmolb.2023.1242059. [20] PENG H, WANG YF, LUO WB. Multifaceted role of branched-chain amino acid metabolism in cancer[J]. Oncogene, 2020, 39( 44): 6747- 6756. DOI: 10.1038/s41388-020-01480-z. [21] BONVINI A, ROGERO MM, COQUEIRO AY, et al. Effects of different branched-chain amino acids supplementation protocols on the inflammatory response of LPS-stimulated RAW 264.7 macrophages[J]. Amino Acids, 2021, 53( 4): 597- 607. DOI: 10.1007/s00726-021-02940-w. [22] LING ZN, JIANG YF, RU JN, et al. Amino acid metabolism in health and disease[J]. Signal Transduct Target Ther, 2023, 8( 1): 345. DOI: 10.1038/s41392-023-01569-3. [23] HU GY, CUI Z, CHEN XY, et al. Suppressing mesenchymal stromal cell ferroptosis via targeting a metabolism-epigenetics axis corrects their poor retention and insufficient healing benefits in the injured liver milieu[J]. Adv Sci(Weinh), 2023, 10( 13): e2206439. DOI: 10.1002/advs.202206439. [24] MOSSMANN D, MÜLLER C, PARK S, et al. Arginine reprograms metabolism in liver cancer via RBM39[J]. Cell, 2023, 186( 23): 5068- 5083. DOI: 10.1016/j.cell.2023.09.011. [25] LÍNDEZ AA MI, REITH W. Arginine-dependent immune responses[J]. Cell Mol Life Sci, 2021, 78( 13): 5303- 5324. DOI: 10.1007/s00018-021-03828-4. [26] SICA A, PORTA C, MORLACCHI S, et al. Origin and functions of tumor-associated myeloid cells(TAMCs)[J]. Cancer Microenviron, 2012, 5( 2): 133- 149. DOI: 10.1007/s12307-011-0091-6. [27] YANG LM, CHU ZL, LIU M, et al. Amino acid metabolism in immune cells: Essential regulators of the effector functions, and promising opportunities to enhance cancer immunotherapy[J]. J Hematol Oncol, 2023, 16( 1): 59. DOI: 10.1186/s13045-023-01453-1. [28] MISSIAEN R, ANDERSON NM, KIM LC, et al. GCN2 inhibition sensitizes arginine-deprived hepatocellular carcinoma cells to senolytic treatment[J]. Cell Metab, 2022, 34( 8): 1151- 1167. DOI: 10.1016/j.cmet.2022.06.010. [29] TRÉZÉGUET V, FATROUNI H, MERCHED AJ. Immuno-metabolic modulation of liver oncogenesis by the tryptophan metabolism[J]. Cells, 2021, 10( 12): 3469. DOI: 10.3390/cells10123469. [30] SOLVAY M, HOLFELDER P, KLAESSENS S, et al. Tryptophan depletion sensitizes the AHR pathway by increasing AHR expression and GCN2/LAT1-mediated kynurenine uptake, and potentiates induction of regulatory T lymphocytes[J]. J Immunother Cancer, 2023, 11( 6): e006728. DOI: 10.1136/jitc-2023-006728. [31] CAMPESATO LF, BUDHU S, TCHAICHA J, et al. Blockade of the AHR restricts a Treg-macrophage suppressive axis induced by L-Kynurenine[J]. Nat Commun, 2020, 11( 1): 4011. DOI: 10.1038/s41467-020-17750-z. [32] DEY S, MONDAL A, DUHADAWAY JB, et al. IDO1 signaling through GCN2 in a subpopulation of gr-1+ cells shifts the IFNγ/IL6 balance to promote neovascularization[J]. Cancer Immunol Res, 2021, 9( 5): 514- 528. DOI: 10.1158/2326-6066.CIR-20-0226. [33] HEZAVEH K, SHINDE RS, KLÖTGEN A, et al. Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress anti-tumor immunity[J]. Immunity, 2022, 55( 2): 324- 340. DOI: 10.1016/j.immuni.2022.01.006. [34] GIRITHAR HN, STAATS PIRES A, AHN SB, et al. Involvement of the kynurenine pathway in breast cancer: Updates on clinical research and trials[J]. Br J Cancer, 2023, 129( 2): 185- 203. DOI: 10.1038/s41416-023-02245-7. [35] LONG G, WANG D, TANG JN, et al. Development of tryptophan metabolism patterns to predict prognosis and immunotherapeutic responses in hepatocellular carcinoma[J]. Aging(Albany NY), 2023, 15( 15): 7593- 7615. DOI: 10.18632/aging.204928. [36] PASCALE RM, PEITTA G, SIMILE MM, et al. Alterations of methionine metabolism as potential targets for the prevention and therapy of hepatocellular carcinoma[J]. Medicina(Kaunas), 2019, 55( 6): 296. DOI: 10.3390/medicina55060296. [37] ROY DG, CHEN J, MAMANE V, et al. Methionine metabolism shapes T helper cell responses through regulation of epigenetic reprogramming[J]. Cell Metab, 2020, 31( 2): 250- 266. DOI: 10.1016/j.cmet.2020.01.006. [38] HUNG MH, LEE JS, MA C, et al. Tumor methionine metabolism drives T-cell exhaustion in hepatocellular carcinoma[J]. Nat Commun, 2021, 12( 1): 1455. DOI: 10.1038/s41467-021-21804-1. [39] SINCLAIR LV, HOWDEN AJ, BRENES A, et al. Antigen receptor control of methionine metabolism in T cells[J]. eLife, 2019, 8: e44210. DOI: 10.7554/eLife.44210. [40] SAINI N, NAAZ A, METUR SP, et al. Methionine uptake via the SLC43A2 transporter is essential for regulatory T-cell survival[J]. Life Sci Alliance, 2022, 5( 12): e202201663. DOI: 10.26508/lsa.202201663. [41] LI JT, YANG H, LEI MZ, et al. Dietary folate drives methionine metabolism to promote cancer development by stabilizing MAT IIA[J]. Signal Transduct Target Ther, 2022, 7( 1): 192. DOI: 10.1038/s41392-022-01017-8. [42] VOGEL A, MEYER T, SAPISOCHIN G, et al. Hepatocellular carcinoma[J]. Lancet, 2022, 400: 1345- 1362. DOI: 10.1016/S0140-6736(22)01200-4. [43] ENDICOTT M, JONES M, HULL J. Amino acid metabolism as a therapeutic target in cancer: A review[J]. Amino Acids, 2021, 53( 8): 1169- 1179. DOI: 10.1007/s00726-021-03052-1. -



 PDF下载 ( 1054 KB)
PDF下载 ( 1054 KB)

 下载:
下载:


