Mechanism of action of circular RNAs in hepatocellular carcinoma
-
摘要: 肝细胞癌(HCC)是全球癌症相关死亡的主要原因之一,存在较高的病死率和复发率,生存率极低。环状RNA (circRNAs)是一类具有组织发育特异性表达模式的单链闭合环状RNA分子,在生物体内发挥调控作用,这些circRNAs通过充当miRNA海绵和靶向蛋白编码基因来调节HCC的进展,但其基本机制仍不清楚。归纳了circRNAs的形成机制及其生物学功能,并综述了circRNAs在HCC发病机制中的作用,提出其作为HCC诊断标志物和治疗靶点的潜在价值。Abstract: Hepatocellular carcinoma( HCC) is one of the leading causes of cancer-related death worldwide and has high mortality and recurrence rates and low survival rates. Circular RNAs( circRNAs) are a class of single-stranded closed circular RNA molecules with tissue-and development-specific expression and play a regulatory role in organisms. These circRNAs regulate the progression of HCC by acting as miRNA sponges and targeted protein-coding genes,but the basic mechanism is still unclear. This article summarizes the formation mechanism and biological functions of circRNAs,reviews the research advances in the role of circRNAs in the pathogenesis of HCC,and points out their potential value as diagnostic markers and therapeutic targets for HCC.
-
Key words:
- carcinoma,hepatocellular /
- circRNAs /
- gene expression regulation
-
检验项目参考区间是临床疾病诊断与健康监测的主要依据,参考区间的准确性、适用性直接影响着临床判断。2012年,我国卫生行业标准发布了血清AST、ALT、GGT、ALP等项目的参考区间,同时指出该参考区间可能受民族、地区影响而致其不适用,各实验室可自行建立参考区间[1]。目前建立参考区间的方法有直接法[2]和间接法[3],直接法需通过建立严格的排除标准而筛选符合要求的参考个体,且此法过程漫长繁琐、难以实施;而间接法只需通过数学统计,凭借快速简便、能得到与直接法相似结果的优势脱颖而出。本研究利用临床实验室信息系统(laboratory information system,LIS)已有数据,用数学统计模型建立AST、ALT、GGT、ALP间接法参考区间,并与国家行业标准比较,以期为临床疾病诊断与健康监测提供依据和参考。
1. 资料与方法
1.1 研究对象
收集本院2019年10月—2020年10月LIS中体检中心健康人群数据,排除信息不全及溶血、脂血、黄疸等不合格样本后,通过比对样本号、姓名、年龄、就诊日期等信息确认为同一来源选择首次测量结果。共入组20~79岁健康人群数据29 443例,其中男性13 423例、女性16 020例。
1.2 仪器与试剂
使用日立公司的7600-210全自动生化分析仪、上海科华公司的试剂与标准品、美国伯乐公司的室内质控品。仪器每年由厂家进行一次校准,每日8点前至少对高、中、低值质控品进行测定(批号:45803,45802,45801)。
1.3 研究方法
AST的检测方法为紫外-苹果酸脱氢酶法,ALT的检测方法为紫外-乳酸脱氢酶法,GGT的检测方法为L-γ-谷氨酰-3-羧基-4-硝基苯氨法,ALP的检测方法为AMP缓冲液法。研究对象空腹8 h后于次日上午7∶ 30—9∶ 30安坐,自肘前静脉真空采血4 ml,常温送至检验科进行上述4种生化指标检测。
1.4 伦理学审查
本研究方案经由吉林大学第一医院伦理委员会批准,批号:2019-249。
1.5 统计学方法
利用SPSS 23.0、Minitab 17、LMS chartmaker Light 2.54、Excel 2016软件分析数据。柯尔莫哥洛夫-斯米诺夫检验判断数据正态性,若其呈偏态分布则使用BOC-COX法转换,此法中的待定变换参数λ由极大似然法求得,其值由所有数据决定。BOX-COX法可针对不同的λ做出不同变换。转换后的数据经P-P图检验为近似正态分布后,使用Turkey法剔除离群值。剔除时把四项指标视为一个整体,若其中任意一个指标符合剔除标准则剔除此人全部信息。Spearman相关分析判断AST、ALT、GGT、ALP与年龄的相关性。LMS法建立连续百分位数曲线。Mann-Whitney U检验比较性别差异,5岁为一年龄组,Z检验比较年龄差异。用数据均值、标准方差及样本量计算Z与Z*值,如Z>Z*则差异有统计学意义,需分组建立参考区间[2]。非参数法计算数据分布的2.5%和97.5%百分位值作为参考区间的上、下限,并用Bootstrap计算其90%置信区间。P < 0.05为差异有统计学意义。
1.6 参考区间的验证
选取本院2020年11月体检中心20~79岁健康人群作为验证个体,对新建立的参考区间进行适用性验证[4],并参照我国行业标准:参考个体多于20例且落在参考区间内的数据≥90%,则通过验证。计算各指标参考区间上、下限值与我国行业标准及其他研究的相对偏差,并与相应参考变化值(reference change value,RCV)比较,其中RCV的计算公式为 (CVa为分析变异系数、CVi为个体内生物学变异系数,通过Westgard网站获得;Z为差异的可能性概率,95%的可能性概率取值1.96),若相对偏差>RCV则认为二者间差异有统计学意义。
2. 结果
2.1 数据分布与离群值剔除
调取的AST、ALT、GGT、ALP数据均不服从正态分布,使用BOX-COX法将其转换为近似正态分布,其中四组数据的待定变换参数λ分别为-0.80、-0.78、0.25、-0.10。使用Turkey法剔除数据2225例,其中男性1213例,女性1012例。正态转换及离群值剔除前后具体数据详见表 1。
表 1 BOX-COX变换及剔除前后数据分布指标 数据类型 样本量(例) 均值 标准差 全距 中位数 P25 P75 AST (U/L) 原始数据 29 443 21.39 9.11 219.70 19.30 16.40 23.60 转换后 29 443 0.09 0.02 0.33 0.09 0.08 0.11 剔除后 27 218 20.58 6.14 40.10 19.20 16.40 23.10 ALT (U/L) 原始数据 29 443 22.69 17.08 385.90 17.80 12.60 26.70 转换后 29 443 0.11 0.05 1.70 0.11 0.08 0.14 剔除后 27 218 21.29 13.01 122.80 17.60 12.70 25.50 GGT (U/L) 原始数据 29 443 25.51 20.39 149.20 18.70 12.50 26.70 转换后 29 443 2.17 0.37 2.59 2.09 1.89 2.37 剔除后 27 218 22.91 14.51 75.80 18.20 12.50 28.90 ALP (U/L) 原始数据 29 443 60.38 17.50 313.90 58.00 48.30 69.70 转换后 29 443 0.66 0.02 0.22 0.66 0.64 0.67 剔除后 27 218 59.94 15.90 92.80 57.90 48.30 69.30 注:剔除部分为离群值。 2.2 AST、ALT、GGT、ALP水平的性别及年龄间比较
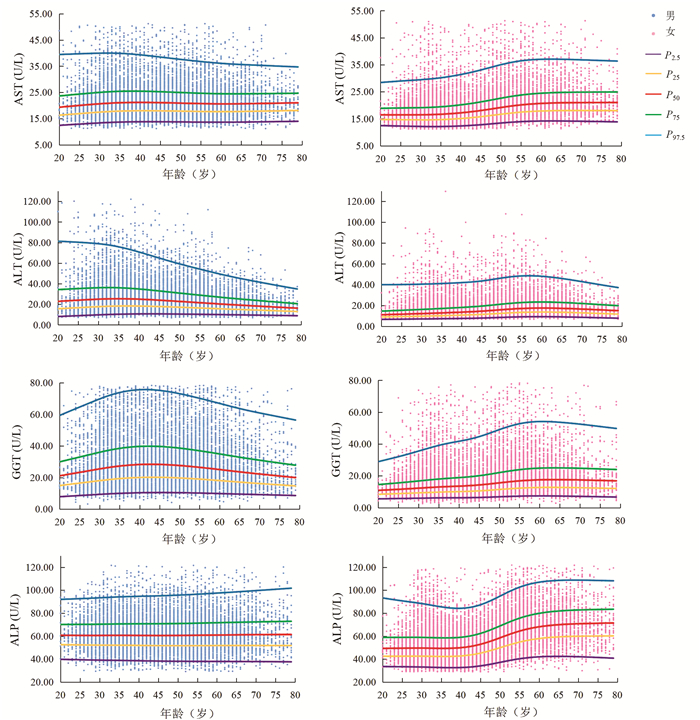
Mann-Whitney U检验结果显示,AST在20~54岁性别间差异有统计学意义(7个年龄段Z值分别为-5.68、-15.59、-30.14、-25.86、-19.22、-15.12、-4.29,P值均 < 0.05)。ALT、GGT、ALP在各年龄组不同性别间差异均有统计学意义(ALT各年龄段Z值分别为-10.27、-24.71、-39.41、-33.86、-28.14、-26.94、-15.17、-11.52、-5.90、-4.77、-3.20、-3.83;GGT各年龄段Z值分别为-12.06、-24.95、-39.70、-34.04、-31.95、-31.61、-23.33、-19.20、-13.54、-9.46、-6.29、-3.65;ALP各年龄段Z值分别为-8.32、-16.36、-22.67、-23.55、-20.42、-14.99、-4.62、-12.77、-10.20、-8.54、-5.03、-2.59,P值均 < 0.05)。Spearman相关结果显示,男性ALT(r=-0.191, P<0.001)、GGT(r=-0.041, P<0.001)与年龄呈负相关;女性AST(r=0.365, P<0.001)、ALT(r=0.310, P<0.001)、GGT(r=0.264, P<0.001)及ALP(r=0.411, P<0.001)与年龄呈正相关。分组后各亚组样本量均大于120例。男性ALT、GGT及女性AST、ALT、GGT、ALP的年龄差异均有统计学意义(Z>Z*)。AST、ALT、GGT、ALP水平连续百分位数曲线见图 1。
2.3 参考区间的建立、验证及比较
用于验证的样本剔除离群值后共10 598例,其中男性6292例、女性4306例。男性、女性共14个亚组,各组验证样本量均>20例,4项生化指标均有>90%的测定值落在本研究参考区间内。使用非参数方法计算的AST、ALT、GGT、ALP参考区间及适用性验证见表 2。为与我国行业标准保持一致,本研究结果取整数。AST、ALT、GGT和女性20~49岁ALP参考区间与我国行业标准的相对偏差均低于RCV,男性及50~79岁女性ALP参考区间与我国行业标准的相对偏差高于RCV。本研究结果与国内外研究结果比较见表 3。
表 2 AST、ALT、GGT、ALP参考区间及适用性验证指标 分组 样本量(例) P2.5 90%置信区间 P97.5 90%置信区间 验证样本量(例) 验证通过率(%) 性别 年龄(岁) 下限 上限 下限 上限 AST (U/L) 男 20~79 12 210 13.50 13.40 13.60 39.30 38.60 39.90 6292 95.12 女 20~49 10 035 12.20 12.10 12.30 32.40 31.61 33.20 2744 95.70 50~79 4973 13.90 13.70 14.00 38.60 37.60 39.60 1562 96.16 ALT (U/L) 男 20~54 8714 10.39 10.10 10.50 70.53 69.13 72.24 4544 93.90 55~79 3496 9.90 9.54 10.00 49.26 47.77 51.47 1748 94.79 女 20~49 10 035 7.40 7.30 7.40 42.60 41.20 43.81 2744 93.95 50~79 4973 9.00 8.84 9.20 49.27 48.03 50.80 1562 94.88 GGT (U/L) 男 20~64 11 195 10.50 10.39 10.80 69.90 69.11 70.50 5732 90.25 65~79 1015 10.00 9.30 10.30 64.20 59.80 66.80 560 94.29 女 20~49 10 035 6.20 6.10 6.30 44.90 43.50 46.59 2744 95.48 50~79 4973 7.40 7.20 7.60 54.23 51.90 58.00 1562 94.30 ALP (U/L) 男 20~79 12 210 37.90 37.60 38.40 95.97 95.02 97.20 6292 92.39 女 20~49 10 035 33.00 32.70 33.30 88.72 87.42 90.40 2744 93.11 50~79 4973 40.20 39.70 41.00 106.30 105.30 107.43 1562 91.87 表 3 本研究与其他研究结果比较指标 本研究(间接法) 我国行业标准(直接法) 俄罗斯(直接法) 沙特阿拉伯(直接法) 我国上海(间接法) RCV(%) 年龄(岁) 男 女 年龄(岁) 男 女 相对偏差(%) 年龄(岁) 男 女 相对偏差(%) 年龄(岁) 男 女 相对偏差(%) 年龄(岁) 男 女 相对偏差(%) AST (U/L) 20~79 14~39 20~19 15~45 13~40 (7.14, 15.38) 18~64 15~41 14~32 (7.14, 5.13) 18~65 11~28 10~24 (21.43, 28.21) 20~79 15~46 13~38 (7.14, 17.95) 38.12 20~49 12~32 (8.33, 25.00) (16.67, 0) (16.67, 25.00) (8.33, 18.75) 38.12 50~79 14~39 (7.14, 2.56) (0, 17.95) (28.57, 38.46) (7.14, 2.56) 38.12 ALT (U/L) 20~54 10~71 20~19 9~60 1~45 (10.00, 15.49) 18~64 11~51 7~31 (10.00, 28.17) 18~65 7~39 5~18 (30.00, 45.07) 20~79 8~555 5~42 (20.00, 22.54) 60.12 55~19 10~49 (10.00, 22.45) (10.00, 4.08) (30.00, 20.41) (20.00, 12.24) 60.12 20~49 7~43 (0, 4.65) (0, 27.91) (28.57, 58.14) (28.57, 7.69) 60.12 50~79 9~49 (22.22, 8.16) (22.22, 36.73) (44.44, 63.27) (44.44, 2.33) 60.12 GGT (U/L) 20~64 11~70 20~19 10~60 1~45 (9.09, 13.m) 18~64 12~69 8~32 (9.09, 0) 18~65 11~65 7~21 (0, 5.80) 20~79 16~52 8~39 (45.45, 25.71) 41.53 65~79 10~64 (0, 6.25) (20.00, 7.81) (10.00, 1.56) (60.00, 18.75) 41.53 20~49 6~45 (16.66, 0) (33.33, 28.89) (16.67, 53.33) (33.33, 13.33) 41.53 50~79 7~54 (0, 16.66) (14.29, 40.74) (0, 61.11) (14.29, 27.78) 41.53 ALP (U/L) 20~79 38~96 20~19 45~125 (18.42, 30.21) 18~64 46~121 (21.05, 26.04) 18~65 39~114 (2.63, 18.75) 19.99 20~49 33~89 20~49 35~100 (6.06, 12.36) 18~44 38~89 (15.15, 0) (18.18, 28.09) 19.99 50~79 40~106 50~19 50~135 (25.00, 27.36) 45~64 44~128 (10.00, 20.75) (2.50, 7.55) 19.99 3. 讨论
血清AST、ALT、GGT、ALP作为常规肝功能检测的四项指标对肝肾疾病的诊断及治疗至关重要。近年有研究表明,AST、ALT与癌细胞代谢密切相关,并与不同类型癌症的预后有关[5];ALT与非酒精性脂肪肝性肝炎的肝脏炎症、肝纤维化及肝脂肪变性程度显著相关[6]。也有研究[7]结果显示,GGT、ALP与肾脏预后独立相关。故建立其准确可靠的参考区间具有重要意义。
本研究中男性随年龄增加,ALT、GGT水平先上升后下降,验证了我国多中心研究[8]中男性ALT、GGT水平在40岁前随年龄升高,在40岁后保持不变或下降的结论。这种趋势的可能原因是,ALT、GGT水平与酒精有关[9],人进入中老年期,中度或重度饮酒人士数量减少。本研究中女性AST、ALT、GGT、ALP水平均随年龄增加,在45~55岁时上升明显,验证了我国多中心研究[8]中女性AST、ALT、GGT、ALP与年龄正相关、我国北京[10]ALP水平在50岁后升高的结论。女性45~55岁呈现出的特殊性可能与其生理变化有关,大多数女性在该年龄段步入更年期、绝经期,性激素波动明显。本研究中男性或女性ALT水平在60岁后均有不同程度下降,验证了美国老年人ALT水平与年龄呈负相关的研究结果[11],提示临床医生在解释患者(尤其是老年人)ALT水平时应考虑年龄因素。
通过相对偏差与RCV比较,本研究自建的男性ALP参考区间上限、女性50~79岁ALP参考区间上、下限与我国行业标准间差异有统计学意义(相对偏差>RCV)。男性GGT参考区间下限与我国上海的研究[12]、女性AST、GGT、ALP参考区间上限与沙特阿拉伯的研究[13]、男性ALP参考区间上、下限及50~79岁女性ALP参考区间上限与俄罗斯的研究[14]差异有统计学意义(相对偏差>RCV)。造成差异的原因可能是:(1)这4项生化指标与BMI有关[14-15],或与人群、种族、遗传、地域、生活习惯、饮食结构等有关;(2)样本量不足,纳入研究的数据不足以代替整个人群;(3)不同实验室建立参考区间的检测方法、试剂、仪器等差异;(4)可能存在异常数据,虽经过离群值剔除但不能确保纳入研究的个体为完全健康没有疾病。自建的ALP参考区间与我国行业标准间的差异可通过增加样本量来改善。另外本研究建立的参考区间通过适用性验证,表明该参考区间适用于本地区人群,证实间接法建立参考区间的可行性。此前本实验室已使用直接法建立了长春地区儿童血清AST、ALT、GGT、ALP参考区间[16-17]。直接法虽是建立参考区间的标准方法,但其过程漫长繁琐,如需面对特殊群体(新生儿、孕妇、老人等)会使数据更加难以收集。若实验室直接引用试剂厂商提供的参考区间,难免存在种族、环境等差异。而间接法利用已有数据既可获得特殊群体足够多的数据量,又无需额外投入,是一种很好的选择,适合不同实验室根据本地区人群、检测系统等实际情况建立参考区间。本研究以LIS数据为基础,运用数学统计模型建立参考区间,虽可能纳入异常数据,但通过适宜数据转换、离群值剔除后计算参考区间可有效弥补这一缺陷[8]。间接法作为一种回顾性研究,其实施过程无法达到与直接法一致的严谨程度,各界对其存在一定争议,故应定期验证间接法建立的参考区间以确保其可靠性。本研究根据数据分布及样本量选择了BOX-COX法、Turkey法与非参数法,目前尚不能评价本研究所用方法的优劣。尽管如此,间接法仍然凭借着简单快速的优势在未来有很好的应用前景。
血清酶与性别、年龄等因素关系密切,故应根据性别及年龄建立参考区间。本研究使用间接法建立了成人血清AST、ALT、GGT、ALP的性别及年龄特异性参考区间。该参考区间与我国行业标准较为一致,证实间接法建立参考区间的可信度和可靠性。本研究为血清酶类参考区间研究提供了基础数据,有益于肝病及其他相关疾病的预防及鉴别诊断。
-
[1] LLOVET JM,ZUCMAN-ROSSI J,PIKARSKY E,et al. Hepatocellular carcinoma[J]. Nat Rev Dis Primers, 2016,2:16018. [2] LEVRERO M. Viral hepatitis and liver cancer:The case of hepatitis C[J]. Oncogene,2006,25(27):3834-3847. [3] DE'ANGELIS N,LANDI F,CARRA MC,et al. Managements of recurrent hepatocellular carcinoma after liver transplantation:A systematic review[J]. World J Gastroenterol,2015,21(39):11185-11198. [4] JECK WR,SORRENTINO JA,WANG K,et al. Circular RNAs are abundant,conserved,and associated with ALU repeats[J]. RNA,2013,19(2):141-157. [5] QIN M,LIU G,HUO X,et al. Hsa_circ_0001649:A circular RNA and potential novel biomarker for hepatocellular carcinoma[J]. Cancer Biomark,2016,16(1):161-169. [6] FU L,WU S,YAO T,et al. Decreased expression of hsa_circ_0003570 in hepatocellular carcinoma and its clinical significance[J]. J Clin Lab Anal,2018,32(2):e22239. [7] HSU MT,COCA-PRADOS M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells[J]. Nature,1979,280(5720):339-340. [8] MENG X,LI X,ZHANG P,et al. Circular RNA:An emerging key player in RNA world[J]. Brief Bioinform,2017,18(4):547-557. [9] LEE Y,RIO DC. Mechanisms and regulation of alternative premRNA splicing[J]. Annu Rev Biochem,2015,84:291-323. [10] ZHANG Y,ZHANG XO,CHEN T,et al. Circular intronic long noncoding RNAs[J]. Mol Cell,2013,51(6):792-806. [11] PIWECKA M,GLAZˇAR P,HERNANDEZ-MIRANDA LR,et al.Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function[J]. Science,2017,357(6357):eaam8526. [12] LIU G,SHI H,DENG L,et al. Circular RNA circ-FOXM1 facilitates cell progression as ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in non-small cell lung cancer[J]. Biochem Biophys Res Commun,2019,513(1):207-212. [13] ZHENG Q,BAO C,GUO W,et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs[J]. Nat Commun,2016,7:11215. [14] MEMCZAK S,JENS M,ELEFSINIOTI A,et al. Circular RNAs are a large class of animal RNAs with regulatory potency[J].Nature,2013,495(7441):333-338. [15] LI Z,HUANG C,BAO C,et al. Corrigendum:Exon-intron circular RNAs regulate transcription in the nucleus[J]. Nat Struct Mol Biol,2017,24(2):194. [16] PAMUDURTI NR,BARTOK O,JENS M,et al. Translation of circRNAs[J]. Mol Cell,2017,66(1):9-21. e7. [17] ZHANG M,ZHAO K,XU X,et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma[J]. Nat Commun,2018,9(1):4475. [18] YANG YB,GAO XY,ZHANG ML,et al. Novel role of FBXW7circular RNA in repressing glioma tumorigenesis[J]. J Natl Cancer Inst,2018,110(3):304-315. [19] YANG Y,FAN XJ,MAO MW,et al. Extensive translation of circular RNAs driven by N-methyladenosine[J]. Cell Res,2017,27(5):626-641. [20] DU WW,ZHANG C,YANG W,et al. Identifying and characterizing circRNA-protein interaction[J]. Theranostics,2017,7(17):4183-4191. [21] DONG W,DAI ZH,LIU FC,et al. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production[J].EBio Medicine,2019,45:155-167. [22] ABDELMOHSEN K,PANDA AC,MUNK R,et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1[J]. RNA Biol,2017,14(3):361-369. [23] ZHANG X,LUO P,JING W,et al. circSMAD2 inhibits the epithelial-mesenchymal transition by targeting miR-629 in hepatocellular carcinoma[J]. Onco Targets Ther,2018,11:2853-2863. [24] MENG J,CHEN S,HAN JX,et al. Twist1 regulates vimentin through Cul2 circular RNA to promote EMT in hepatocellular carcinoma[J]. Cancer Res,2018,78(15):4150-4162. [25] LIANG WC,WONG CW,LIANG PP,et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway[J]. Genome Biol,2019,20(1):84. [26] SUN S,WANG W,LUO X,et al. Circular RNA circ-ADD3 inhibits hepatocellular carcinoma metastasis through facilitating EZH2 degradation via CDK1-mediated ubiquitination[J]. Am J Cancer Res,2019,9(8):1695-1707. [27] GUO J,DUAN H,LI Y,et al. A novel circular RNA circ-ZNF652promotes hepatocellular carcinoma metastasis through inducing snail-mediated epithelial-mesenchymal transition by sponging miR-203/miR-502-5p[J]. Biochem Biophys Res Commun,2019,513(4):812-819. [28] FU L,CHEN Q,YAO T,et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma[J]. Oncotarget,2017,8(27):43878-43888. [29] HAN D,LI J,WANG H,et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression[J]. Hepatology,2017,66(4):1151-1164. [30] XU L,FENG X,HAO X,et al. CircSETD3(Hsa_circ_0000567)acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth[J]. J Exp Clin Cancer Res,2019,38(1):98. [31] HUANG XY,HUANG ZL,HUANG J,et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis[J]. J Exp Clin Cancer Res,2020,39(1):20. [32] ZHAI Z,FU Q,LIU C,et al. Emerging roles Of hsa-circ-0046600 targeting the miR-640/HIF-1αsignalling pathway in the progression of HCC[J]. Onco Targets Ther,2019,12:9291-9302. [33] LUO Y,FU Y,HUANG R,et al. CircRNA_101505 sensitizes hepatocellular carcinoma cells to cisplatin by sponging miR-103 and promotes oxidored-nitro domain-containing protein1 expression[J]. Cell Death Discov,2019,5:121. [34] LI J,QIN X,WU R,et al. Circular RNA circFBXO11 modulates hepatocellular carcinoma progress and oxaliplatin resistance through miR-605/FOXO3/ABCB1 axis[J]. J Cell Mol Med,2020,24(9):5152-5161. [35] CHEN H,LIU S,LI M,et al. circ_0003418 inhibits tumorigenesis and cisplatin chemoresistance through Wnt/β-catenin pathway in hepatocellular carcinoma[J]. Onco Targets Ther,2019,12:9539-9549. [36] ZHU K,ZHAN H,PENG Y,et al. Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis B virus-related hepatocellular carcinoma[J]. Carcinogenesis,2020,41(3):296-302. [37] CHEN D,ZHANG C,LIN J,et al. Screening differential circular RNA expression profiles reveal that hsa_circ_0128298 is a biomarker in the diagnosis and prognosis of hepatocellular carcinoma[J]. Cancer Manag Res,2018,10:1275-1283. [38] QU S,YANG X,LI X,et al. Circular RNA:A new star of noncoding RNAs[J]. Cancer Lett,2015,365(2):141-148. [39] DENG X,CAO M,ZHANG J,et al. Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer[J]. Biomaterials,2014,35(14):4333-4344. [40] TATOMER DC,LIANG D,WILUSZ JE. Inducible expression of eukaryotic circular RNAs from plasmids[J]. Methods Mol Biol,2017,1648:143-154. 期刊类型引用(3)
1. 何冰,王艺婷,李雪文,许建成. 长春市不同年龄段健康成人血清心肌酶谱水平观察. 检验医学与临床. 2024(02): 151-155 .  百度学术
百度学术2. 孔焱,李晓慧,王辉,李岩,孙悦,苗强. 间接法建立新疆克拉玛依地区常规肝功能和血脂生化项目参考区间. 国际检验医学杂志. 2024(07): 858-861+866 .  百度学术
百度学术3. 张春娇,黄蓉,蔡婷婷,顾进. 基于间接法建立肾功能检验项目的生物参考区间. 湖南师范大学学报(医学版). 2024(01): 43-47 .  百度学术
百度学术其他类型引用(1)
-




 PDF下载 ( 420 KB)
PDF下载 ( 420 KB)

 下载:
下载:

 百度学术
百度学术
 下载:
下载: