微RNA风险评分模型预测肝细胞癌预后的价值分析
DOI: 10.3969/j.issn.1001-5256.2021.05.026
Value of a microRNA risk score model in predicting the prognosis of hepatocellular carcinoma
-
摘要:
目的 下载癌症基因组图谱(TCGA)数据库中肝细胞癌(HCC)miRNA转录组数据进行数据挖掘,筛选与HCC预后相关的miRNA,构建miRNA风险评分模型,并评估其对HCC预后的预测价值。 方法 从TCGA数据库下载HCC样本miRNA表达量及临床数据,应用R语言筛选出HCC与癌旁组织的差异表达miRNA,将其与临床数据整合后,随机分为训练集和测试集,对训练集依次进行单因素Cox及LASSO-Cox回归分析,筛选与HCC预后相关的miRNA并构建风险评分模型,并使用Kaplan-Meier分析评估模型的稳健性及是否可以预测同一临床分期的患者预后,最后通过受试者工作特征曲线(ROC)计算曲线下面积(AUC),在训练集、测试集及二者的合集中比较该模型和传统TNM分期的预测准确性。 结果 共筛选出300个差异基因,LASSO-Cox回归分析显示hsa-miR-139-5p、hsa-miR-1180-3p、hsa-miR-1269b、hsa-miR-3680-3p、hsa-miR-509-3-5p、hsa-miR-31-5p与HCC预后相关,根据构建的miRNA风险评分模型,计算每个样本的风险得分,并根据中位风险得分值将样本划分为高风险组和低风险组。Kaplan-Meier曲线显示,训练集与测试集的高风险组患者的生存率显著低于低风险组患者(P<0.05)。ROC曲线结果显示,训练集、测试集及合集样本中,miRNA模型及TNM分期的AUC分别为0.817、0.667,0.808、0.665与0.814、0.663。独立预后分析结果显示,该miRNA评分模型可作为HCC的独立预后因子(P<0.05)。 结论 hsa-miR-139-5p、hsa-miR-1180-3p、hsa-miR-1269b、hsa-miR-3680-3p、hsa-miR-509-3-5p、hsa-miR-31-5p与HCC预后相关,在训练集、测试集及合集样本中miRNA风险评分模型的预测准确性均优于TNM分期。分层分析表明,该模型还可预测同一TNM分期患者的预后,在临床工作中具有一定参考价值,可作为独立预测HCC患者预后的模型。 Abstract:Objective To screen out the microRNAs (miRNAs) associated with the prognosis of hepatocellular carcinoma (HCC) through data mining of miRNA transcriptome data of HCC downloaded from The Cancer Genome Atlas (TCGA) database, to establish a miRNA risk score model, and to investigate its value in predicting the prognosis of HCC. Methods The miRNA expression data and clinical data of HCC samples were downloaded from TCGA database and R language was used to screen out differentially expressed miRNAs between HCC tissue and adjacent tissue, which were randomly divided into training set and testing set after being integrated into clinical data. Univariate Cox regression analysis and least absolute shrinkage and selection operator (LASSO) Cox regression analysis were performed for the training set to screen out the miRNAs associated with the prognosis of HCC, and then a miRNA risk score model was established. The Kaplan-Meier method was used to evaluate the robustness of the model and whether it could predict the prognosis of patients in the same clinical stage. Finally, the receiver operating characteristic (ROC) curve was plotted and the area under the ROC curve (AUC) was calculated to compare the predictive accuracy of the model versus TNM staging in the training set, the testing set, and the entire set. Results A total of 300 differentially expressed miRNAs were screened out and the LASSO Cox regression analysis revealed that hsa-miR-139-5p, hsa-miR-1180-3p, hsa-miR-1269b, hsa-miR-3680-3p, hsa-miR-509-3-5p, and hsa-miR-31-5p were associated with the prognosis of HCC. The risk score was calculated for each sample according to the established miRNA risk score model, and the samples were divided into high-risk group and low-risk group according to the median risk score. The Kaplan-Meier curve showed that in both training and testing sets, the high-risk group had a significantly lower survival rate than the low-risk group (P < 0.05). The ROC curve was used to evaluate the prediction efficiency of this model, and the results showed that in the training set, the testing set, and the entire set, the miRNA model had an AUC of 0.817, 0.808, and 0.814, respectively, while TNM staging had an AUC of 0.667, 0.665, and 0.663, respectively. The results of independent prognostic analysis also showed that this miRNA score model could be used as an independent prognostic factor for HCC (P < 0.05). Conclusion Hsa-miR-139-5p, hsa-miR-1180-3p, hsa-miR-1269b, hsa-miR-3680-3p, hsa-miR-509-3-5p, and hsa-miR-31-5p are associated with the prognosis of HCC, and the miRNA risk score model has a better prediction accuracy than TNM staging in the training set, the testing set, and the entire set. The stratified analysis also shows that the model can predict the prognosis of patients within the same TNM stage, and therefore, it has a certain reference value in clinical practice and can be used as an independent model for predicting the prognosis of HCC patients. -
Key words:
- Hepatocellular Carcinoma /
- MicroRNAs /
- Prognosis
-
肝细胞癌(HCC)是最常见的原发性肝癌,其发病率在所有恶性肿瘤中排第6位,致死率排第4位,世界卫生组织估计,截至2030年,全球每年将有超过100万死于HCC的患者[1-2]。由于HCC起病隐匿,进展迅速且异质性高,尽管外科手术、射频消融、化学栓塞甚至肝移植已广泛用于HCC治疗,但HCC患者的存活率仍然较低[3-4]。由于与HCC发生和发展有关的生物学过程非常复杂,迄今为止尚无敏感高效的预后生物标志物,因此,有必要探索新的能够高效预测HCC预后的生物标志物或模型,用于HCC的诊断、预后和治疗。
微RNA(microRNA, miRNA)是一类参与mRNA转录后调控的非编码小RNA,通过与目标mRNA的3′非编码区结合,抑制转译或降解mRNA,继而在转录后水平调控靶基因的表达,从而影响细胞生物学功能,miRNA的差异表达在多种恶性肿瘤的发生发展中发挥关键作用[5-6],而且多项研究[7-9]已经确定miRNA模型能够预测恶性肿瘤患者的预后,比如乳腺癌、前列腺癌及卵巢癌。而在本次研究中,首先从TCGA中下载HCC基因及临床数据,随后找出与HCC预后相关的miRNA,构建预测HCC预后的miRNA风险评分模型,并验证其检验效能。
1. 资料与方法
1.1 数据下载与整理
从TCGA数据库(https://portal.gdc.cancer.gov/)中下载HCC miRNA转录组数据及临床数据。
1.2 差异分析
利用R语言的limma包对HCC与正常肝组织之间的miRNA进行基因差异表达对比分析,并设置差异倍数log2FC(fold change,FC)>1.0且P<0.01。
1.3 miRNA风险评分模型的构建
从临床数据中提取生存信息,包括生存时间和生存状态,与差异表达的miRNA进行整合,利用R语言caret包将其随机分成训练集和测试集,利用survival、glmnet、survminer包对训练集进行单因素Cox回归分析,并设定P<0.01为差异具有统计学意义。然后通过10倍交叉验证的LASSO-Cox回归分析进一步筛选预后miRNA,并计算每个miRNA的危险比(HR)和回归系数,基于miRNA表达水平及回归系数的线性组合构建与HCC预后相关的风险评分模型。
1.4 miRNA风险评分模型的验证
根据风险评分模型计算出训练集中每个样本的风险得分,并根据中位风险得分值将样本分为高风险组和低风险组,采用log-rank检验的Kaplan-Meier生存曲线评估高风险组与低风险组的预后差异,组间差异比较时设定P<0.05为差异具有统计学意义,同时在测试集中评估其稳健性。为了评估该模型是否可以预测同一临床分期的患者预后,对不同TNM分期的患者进行了分层分析,将HCC患者分为低TNM组(Ⅰ和Ⅱ期)和高TNM组(Ⅲ和Ⅳ期),同样采用log-rank检验的Kaplan-Meier生存曲线评估高TNM组和低TNM组的预后差异,并设定P<0.05为差异具有统计学意义。
1.5 miRNA风险评分模型的评估
通过R语言的survival ROC包绘制ROC曲线,计算出ROC曲线下面积(AUC),并与传统TNM分期比较,评估该模型的预测准确性,其中AUC>0.7被认为具有良好的预测效能。最后对风险评分模型和临床特征进行单因素和多因素Cox回归分析评估该模型预后独立性,并设定P<0.05为差异具有统计学意义。
2. 结果
2.1 一般资料
miRNA样本共425例,包括375例HCC样本及50例癌旁样本,并将临床数据中生存时间、生存状态及TNM分期缺失的数据删除,获得临床数据352例(表 1)。
表 1 TCGA数据库HCC患者的临床数据项目 数值[例(%)] 生存状态 存活 234(66.5) 死亡 118(33.5) 性别 女性 112(31.8) 男性 240(68.2) 年龄 <60岁 166(47.2) ≥60 186(52.8) 肿瘤分级 G1 46(13.1) G2 171(48.6) G3 120(34.1) G4 13(3.7) 未知 2(0.5) TNM分期 Ⅰ 175(49.7) Ⅱ 86(24.4) Ⅲ 86(24.4) Ⅳ 5(1.5) T分期 T1 176(50.0) T2 88(25.0) T3 77(21.9) T4 10(2.8) 未知 1(0.3) M分期 M0 269(76.4) M1 4(1.2) 未知 79(22.4) N分期 N0 257(73.0) N1 4(1.2) 未知 91(25.8) 2.2 差异表达的miRNA
利用R语言对HCC及癌旁组织进行miRNA差异表达分析,以log2FC>1.0且P<0.01为标准,共筛选出300个差异表达miRNAs,上调基因260个,下调基因40个。
2.3 miRNA风险评分模型的构建
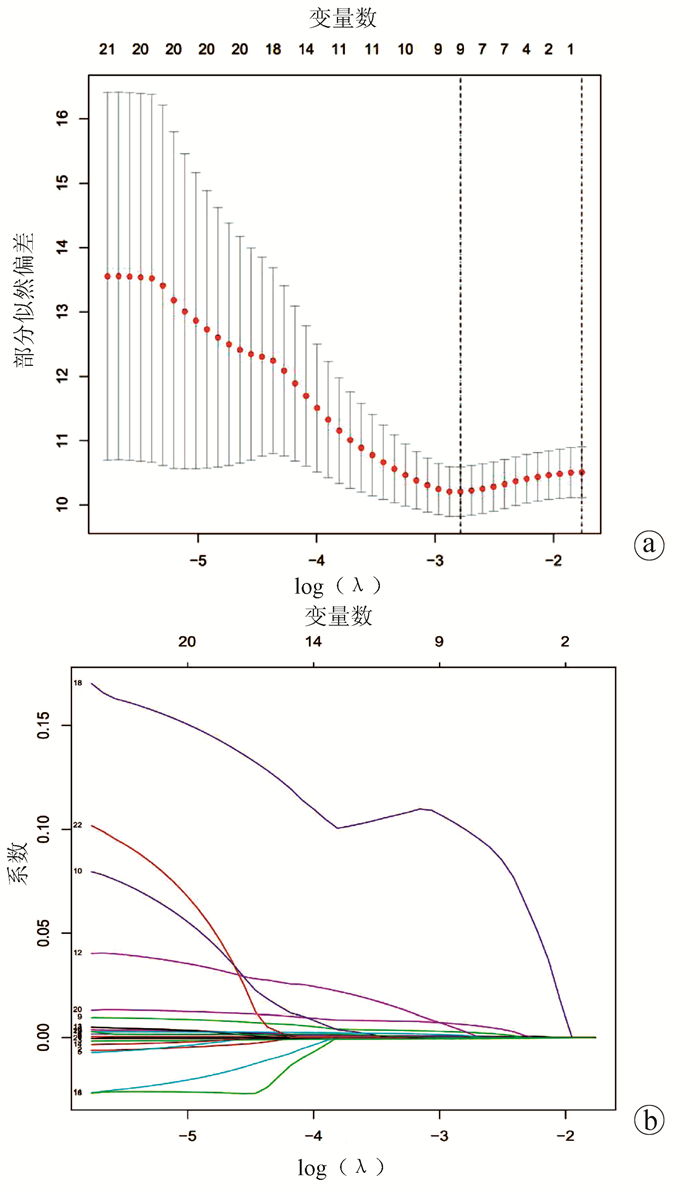
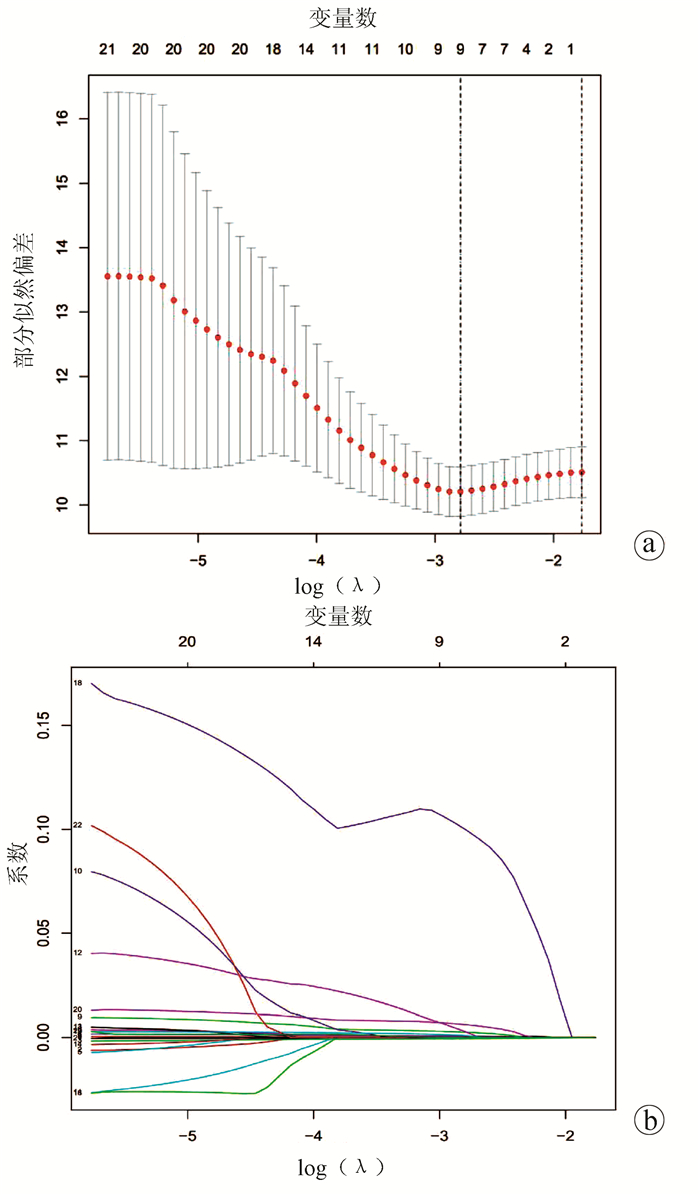
将差异表达的miRNA与临床数据进行整合匹配,共得到347个样本,利用R语言将其按照1∶ 1随机分成训练集(n=175)和测试集(n=172),对训练集依次进行单因素Cox及10倍交叉验证的LASSO-Cox回归分析。单因素Cox回归分析显示,23个miRNAs与HCC预后相关(P<0.01)。然后通过LASSO回归进一步筛选miRNA,结果显示9个miRNAs: hsa-miR-139-5p、hsa-miR-1180-3p、hsa-miR-4652-5p、hsa-miR-1269b、hsa-miR-122b-5p、hsa-miR-3677-5p、hsa-miR-3680-3p、hsa-miR-509-3-5p、hsa-miR-31-5p与HCC预后显著相关(图 1)。
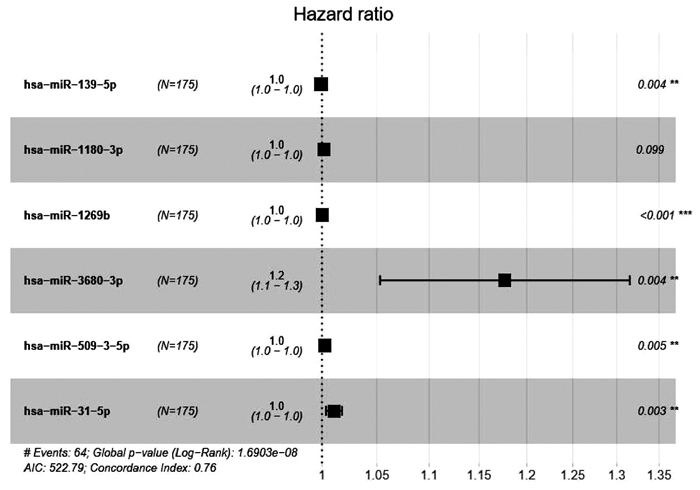
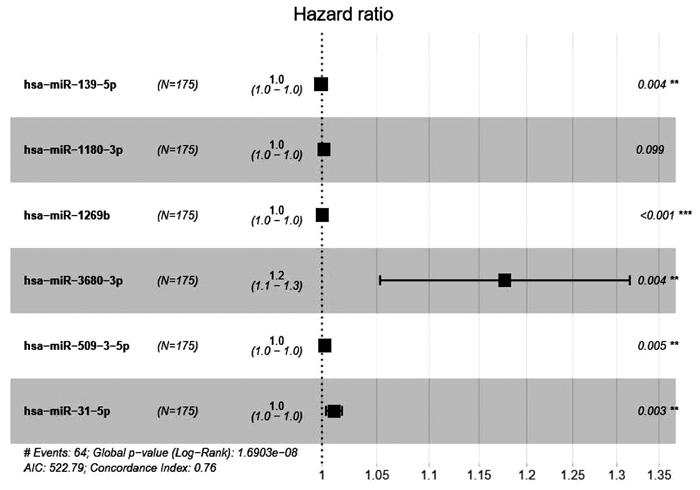
最后将LASSO回归筛选出的miRNAs拟合到多因素Cox回归分析,最终确定了包括hsa-miR-139-5p、hsa-miR-1180-3p、hsa-miR-1269b、hsa-miR-3680-3p、hsa- miR-509-3-5p、hsa-miR-31-5p在内的6个miRNAs,构建风险评分公式并绘制森林图(图 2)。风险评分公式:hsa-miR-139- 5p×(-0.001 07)+hsa-miR-1180-3p×0.001 57+hsa-miR-1269b×0.000 26+hsa-miR-3680-3p×0.162 79 +hsa-miR-509-3-5p×0.002 46+hsa-miR-31-5p×0.010 67,计算每个样品的风险得分。在这些miRNAs中,hsa-miR-1180-3p、hsa-miR-1269b、hsa-miR-3680-3p、hsa-miR-509-3-5p、hsa-miR-31-5p的系数为正,表明这些基因表达水平越高,生存期越短,而hsa-miR-139-5p的系数为负,表明该基因表达水平越高,生存期越长。
2.4 miRNA风险评分模型的验证
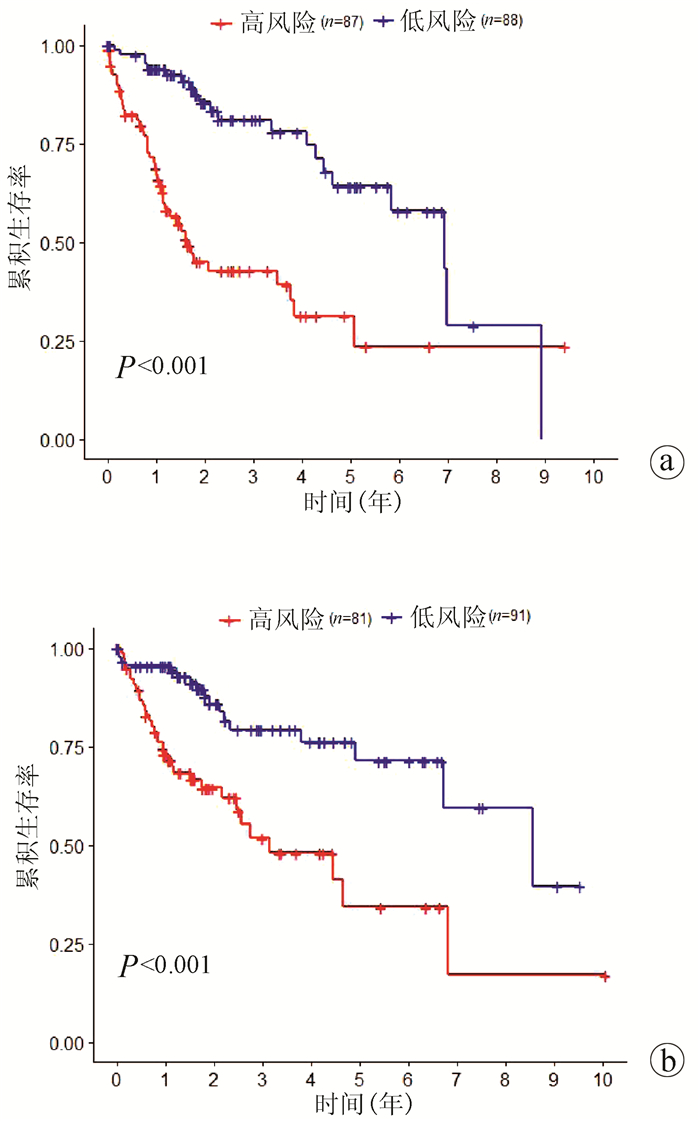
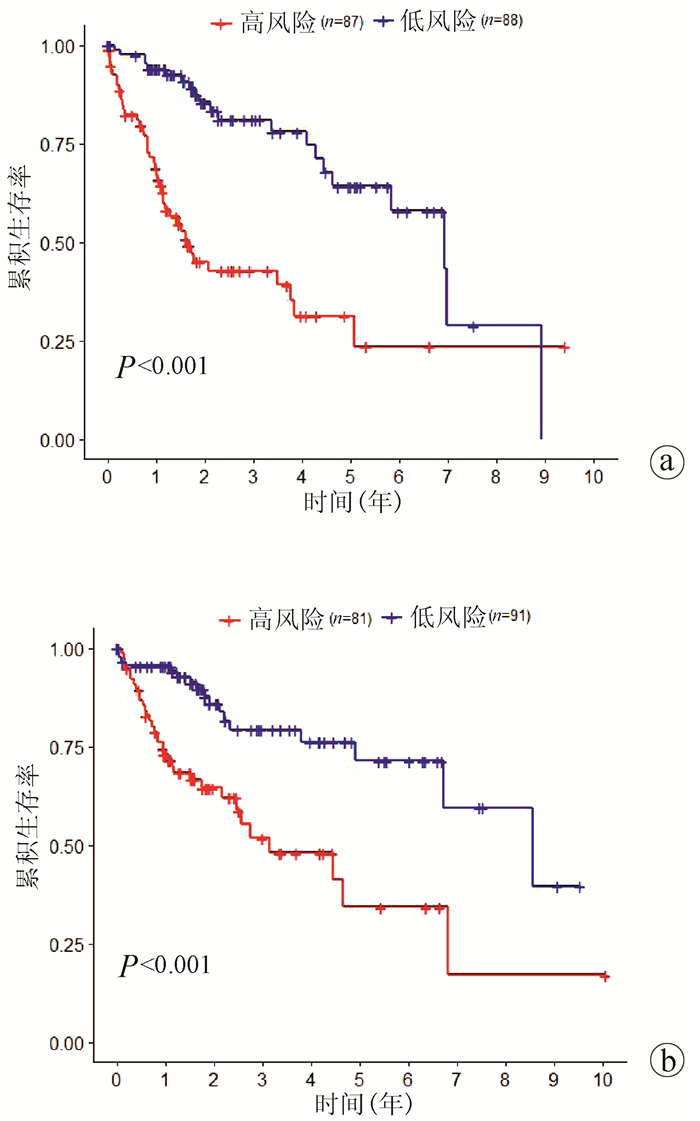
根据miRNA风险评分模型,计算训练集中每个样本的风险得分,再根据中位风险得分值(n=0.918 637 4)将训练集分为高风险组(n=87)和低风险组(n=88)。生存曲线显示(图 3a)高风险组患者的生存率明显低于低风险组,差异具有统计学意义(P<0.05)。同样地,根据相同的风险评分模型及中位风险得分值(n=0.918 637 4),将测试集中的患者分为高风险组(n=81)和低风险组(n=91),生存曲线显示(图 3b),高风险组患者的生存率明显低于低风险组,差异具有统计学意义(P<0.05)。
随后根据样本的风险得分排名,分别绘制训练集和测试集的风险图、生存状态图及与HCC预后相关的6个miRNAs的风险热图(图 4、5)。结果表明高风险评分的患者预后较差,且系数为正的miRNAs(hsa-miR-1180-3p、hsa-miR-1269b、hsa-miR-3 680-3p、hsa-miR-509-3-5p、hsa-miR-31-5p)表达上调,而低风险评分的患者预后较好,且系数为负的miRNA(hsa-miR-139-5p)表达上调。
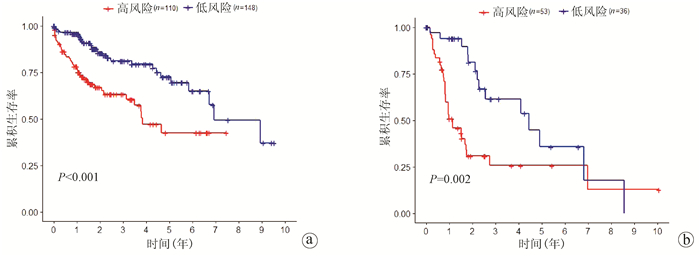
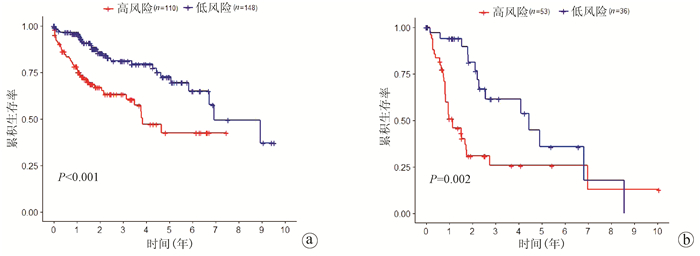
将患者按TNM分期分为低TNM组(Ⅰ和Ⅱ期)和高TNM组(Ⅲ和Ⅳ期),Kaplan-Meier曲线显示低TNM组、高TNM组的高风险患者的生存率显著低于低风险患者(P<0.05),表明该模型可以预测同一临床分期的患者预后。
2.5 miRNA风险评分模型的评估
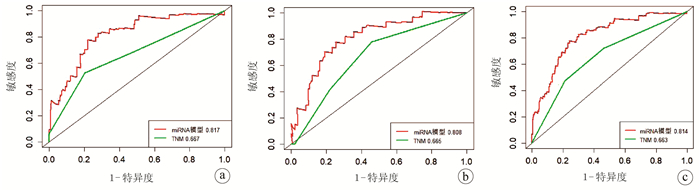
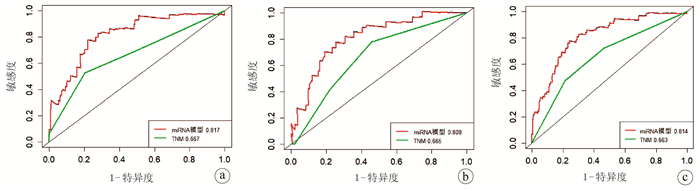
利用R语言绘制miRNA模型及TNM分期的ROC曲线并计算AUC,结果显示训练集(图 7a)中,miRNA模型及TNM分期的AUC分别为0.817、0.667,测试集(图 7b)分别为0.808、0.665,及合集样本(图 7c)分别为0.814、0.663,miRNA模型预测准确性均优于TNM分期。
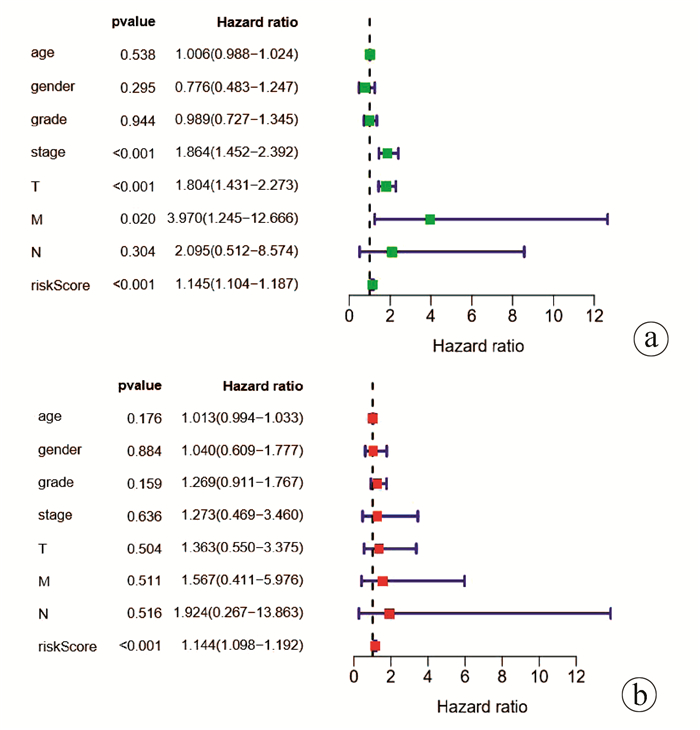
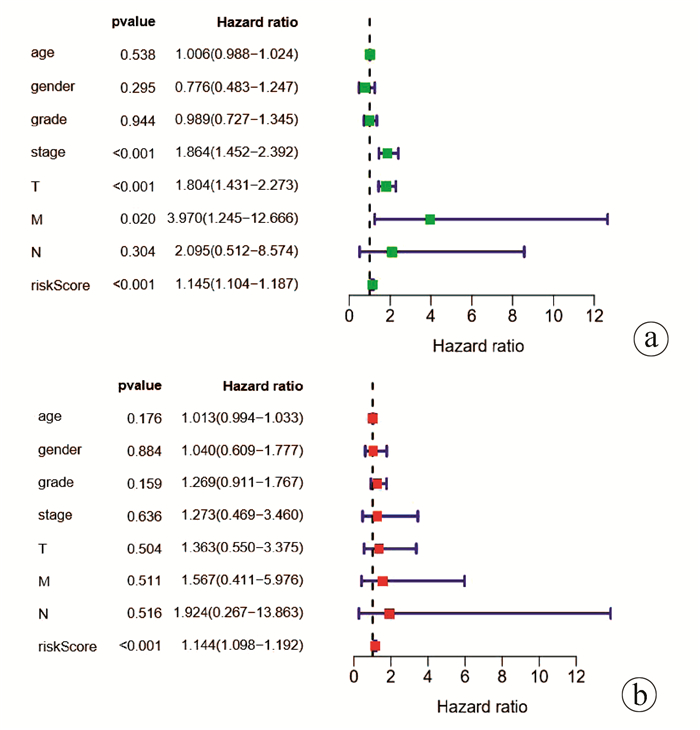
将年龄、性别、肿瘤分级、TNM分期、miRNA评分分别进行单因素、多因素Cox独立预后分析,单因素分析结果显示T分期、M分期、TNM分期及miRNA评分是HCC患者预后的相关因素(P值均<0.05)(图 8a),多因素分析结果显示miRNA评分模型可作为HCC的独立预后因子(P<0.05)(图 8b)。
3. 讨论
原发性肝癌是常见的恶性肿瘤,在各个年龄段均可发病,发病率和死亡率比较高,预后很差。目前,用于预测HCC预后主要依据传统的TNM分期[10],尽管TNM分期在恶性肿瘤的诊断和治疗中起着重要作用,但由于肿瘤的异质性和个体差异,其无法反映内部的生物学过程和疾病进展,因此有必要寻找一种新的能够高效预测HCC预后的生物标志物或模型,辅助临床工作。
在本次研究中,首先从TCGA中下载HCC的miRNA表达量及临床数据,TCGA是由美国国立卫生研究院支持发起的,包括30多种癌症,旨在提供不同癌症的全面基因分析并建立与临床结果的相关性,这些数据对于癌症的研究具有巨大的潜力,并且已经利用这些数据取得了许多成果[11-16]。当作者试图从训练集中确定预后特征时,由于样本量小,基因数量众多,因此对训练集中的数据进行了10倍交叉验证的LASSO-Cox回归分析。其中LASSO是一种创新的回归变量选择方法,在系数的绝对值之和小于一个常数的情况下将残差平方和最小化,从而使某些回归系数严格等于0,从而选择出对因变量影响较大的自变量,是通过构造一个惩罚函数得到一个较为精炼的模型[17],从而减少模型的过度拟合。而交叉验证法是比较常用的推测估计惩罚系数λ的方法,而λ值最终是由使平均交叉验证误差最小的最小化λ确定的。然后根据Cox多因素回归分析,得出和HCC预后相关的6种miRNAs。通过回顾已发表的文献[18-20],作者发现miRNA的失控与HCC的发生、耐药、预后相关,广泛参与HCC中肿瘤抑制基因的失活和癌基因的激活。其中hsa-miR-1180-3p、hsa-miR-1269b、hsa-miR-3680-3p、hsa-miR-509-3-5p、hsa-miR-31-5p的表达在HCC组织中上调,可能起致癌作用,hsa-miR-139-5p在HCC组织中的表达下调,可能起抑癌作用,而hsa-miR-139-5p可以通过减少SLITRK4的表达抑制HCC细胞的生长[21]。然后基于回归系数构建了风险评分模型,计算出训练集中每位患者的风险得分,然后根据中位风险得分值将其分为高风险组和低风险组,并在测试集中验证了该miRNA风险评分模型的预后价值。生存曲线分析表明,训练集、测试集的高风险组和低风险组的生存曲线均观察到明显的分离(P值均<0.05)。为了检验该模型预测效能,绘制了ROC曲线,并计算了相应的AUC,将其与传统TNM分期比较,结果显示在训练集、测试集及合集样本中miRNA模型的预测准确性均优于TNM分期。最后将年龄、性别、分级、分期、miRNA评分模型分别进行单因素、多因素Cox独立预后分析,结果显示该miRNA评分模型可作为HCC的独立预后因子。该模型与传统TNM分期相比较,能够更好地反应内在生物学进程和疾病进展,受主观因素影响少,且该预测模型具有可评估性和可重复性等优点,可与传统TNM分期互补,共同应用于临床工作。同时本研究也有一定的不足,所有分析均通过TCGA数据库完成,以后可同时设置内部验证集和外部验证集(如GEO数据库)验证该模型预测准确性,而且未进行相关实验,这些都有待进一步研究工作。
-
表 1 TCGA数据库HCC患者的临床数据
项目 数值[例(%)] 生存状态 存活 234(66.5) 死亡 118(33.5) 性别 女性 112(31.8) 男性 240(68.2) 年龄 <60岁 166(47.2) ≥60 186(52.8) 肿瘤分级 G1 46(13.1) G2 171(48.6) G3 120(34.1) G4 13(3.7) 未知 2(0.5) TNM分期 Ⅰ 175(49.7) Ⅱ 86(24.4) Ⅲ 86(24.4) Ⅳ 5(1.5) T分期 T1 176(50.0) T2 88(25.0) T3 77(21.9) T4 10(2.8) 未知 1(0.3) M分期 M0 269(76.4) M1 4(1.2) 未知 79(22.4) N分期 N0 257(73.0) N1 4(1.2) 未知 91(25.8) -
[1] FORNER A, REIG M, BRUIX J. Hepatocellular carcinoma[J]. Lancet, 2018, 391(10127): 1301-1314. DOI: 10.1016/S0140-6736(18)30010-2. [2] VILLANUEVA A. Hepatocellular carcinoma[J]. New Engl J Med, 2019, 380(15): 1450-1462. DOI: 10.1056/NEJMra1713263. [3] KULIK L, EL-SERAG HB. Epidemiology and management of hepatocellular carcinoma[J]. Gastroenterology, 2019, 156(2): 477-491. e1. DOI: 10.1053/j.gastro.2018.08.065. [4] YANG JD, HAINAUT P, GORES GJ, et al. A global view of hepatocellular carcinoma: Trends, risk, prevention and management[J]. Nat Rev Gastroenterol Hepatol, 2019, 16(10): 589-604. DOI: 10.1038/s41575-019-0186-y. [5] LIN S, GREGORY RI. MicroRNA biogenesis pathways in cancer[J]. Nat Rev Cancer, 2015, 15(6): 321-333. DOI: 10.1038/nrc3932. [6] RUPAIMOOLE R, SLACK FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases[J]. Nat Rev Drug Discov, 2017, 16(3): 203-222. DOI: 10.1038/nrd.2016.246. [7] YERUKALA SATHIPATI S, HO SY. Identifying a miRNA signature for predicting the stage of breast cancer[J]. Sci Rep, 2018, 8(1): 16138. DOI: 10.1038/s41598-018-34604-3. [8] KANWAL R, PLAGA AR, LIU X, et al. MicroRNAs in prostate cancer: Functional role as biomarkers[J]. Cancer Lett, 2017, 407: 9-20. DOI: 10.1016/j.canlet.2017.08.011. [9] DEB B, UDDIN A, CHAKRABORTY S. miRNAs and ovarian cancer: An overview[J]. J Cell Physiol, 2018, 233(5): 3846-3854. DOI: 10.1002/jcp.26095. [10] Bureau of Medical Administration National Health Commission of the People's Republic of China. Guidelines for diagnosis and treatment of primary liver cancer in China (2019 edition)[J]. J Clin Hepatol, 2020, 36(2): 277-292. DOI: 10.3969/j.issn.1001-5256.2020.02.007.中华人民共和国国家卫生健康委员会医政医管局. 原发性肝癌诊疗规范(2019年版)[J]. 临床肝胆病杂志, 2020, 36(2): 277-292. DOI: 10.3969/j.issn.1001-5256.2020.02.007. [11] TOMCZAK K, CZERWIŃSKA P, WIZNEROWICZ M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge[J]. Contemp Oncol (Pozn), 2015, 19(1A): a68-a77. DOI: 10.5114/wo.2014.47136. [12] LONG J, ZHANG L, WAN X, et al. A four-gene-based prognostic model predicts overall survival in patients with hepatocellular carcinoma[J]. J Cell Mol Med, 2018, 22(12): 5928-5938. DOI: 10.1111/jcmm.13863. [13] WANG X, GAO J, ZHOU B, et al. Identification of prognostic markers for hepatocellular carcinoma based on miRNA expression profiles[J]. Life Sci, 2019, 232: 116596. DOI: 10.1016/j.lfs.2019.116596. [14] JIA D, LI S, LI D, et al. Mining TCGA database for genes of prognostic value in glioblastoma microenvironment[J]. Aging (Albany NY), 2018, 10(4): 592-605. DOI: 10.18632/aging.101415. [15] LI Y, GU J, XU F, et al. Transcriptomic and functional network features of lung squamous cell carcinoma through integrative analysis of GEO and TCGA data[J]. Sci Rep, 2018, 8(1): 15834. DOI: 10.1038/s41598-018-34160-w. [16] NAGY A, LANCZKY A, MENYHART O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets[J]. Sci Rep, 2018, 8(1): 9227. DOI: 10.1038/s41598-018-27521-y. [17] TIBSHIRANI R. The lasso method for variable selection in the Cox model[J]. Stat Med, 1997, 16(4): 385-395. DOI: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [18] WEI L, WANG X, LV L, et al. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma[J]. Mol Cancer, 2019, 18(1): 147. DOI: 10.1186/s12943-019-1086-z. [19] SADRI NAHAND J, BOKHARAEI-SALIM F, SALMANINEJAD A, et al. microRNAs: Key players in virus-associated hepatocellular carcinoma[J]. J Cell Physiol, 2019, 234(8): 12188-12225. DOI: 10.1002/jcp.27956. [20] WONG CM, TSANG FH, NG IO. Non-coding RNAs in hepatocellular carcinoma: Molecular functions and pathological implications[J]. Nat Rev Gastroenterol Hepatol, 2018, 15(3): 137-151. DOI: 10.1038/nrgastro.2017.169. [21] HUA S, LEI L, DENG L, et al. miR-139-5p inhibits aerobic glycolysis, cell proliferation, migration, and invasion in hepatocellular carcinoma via a reciprocal regulatory interaction with ETS1[J]. Oncogene, 2018, 37(12): 1624-1636. DOI: 10.1038/s41388-017-0057-3. 期刊类型引用(1)
1. 王凤松,朱刘洋,白易,张雅敏. 基于肿瘤突变负荷构建肝细胞癌风险评分预后模型. 天津医科大学学报. 2022(01): 20-26 .  百度学术
百度学术其他类型引用(0)
-
 微RNA风险评分模型预测肝细胞癌预后的价值分析 图4.pdf
微RNA风险评分模型预测肝细胞癌预后的价值分析 图4.pdf

 微RNA风险评分模型预测肝细胞癌预后的价值分析 图5.pdf
微RNA风险评分模型预测肝细胞癌预后的价值分析 图5.pdf

-




 PDF下载 ( 4272 KB)
PDF下载 ( 4272 KB)

 下载:
下载:








 下载:
下载:


 百度学术
百度学术