Gilbert综合征与Crigler-Najjar综合征Ⅱ型患者UGT1A1基因多态性分析
DOI: 10.3969/j.issn.1001-5256.2022.02.026
UGT1A1 gene polymorphisms in patients with Gilbert syndrome and Crigler-Najjar syndrome type Ⅱ
-
摘要:
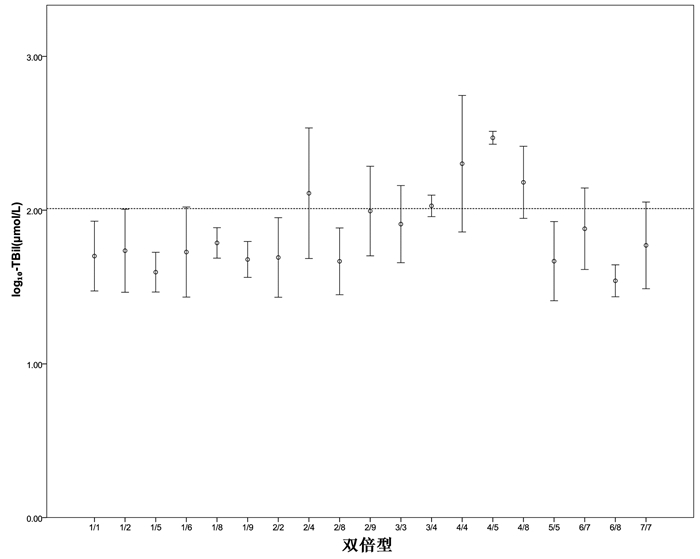
目的 分析Gilbert综合征(GS)与Crigler-Najjar综合征Ⅱ型(CN-2)患者UGT1A1基因突变位点、单倍型及双倍型的差异。 方法 回顾性分析2010年1月1日—2019年12月31日就诊于首都医科大学附属北京佑安医院的138例GS与CN-2患者临床资料,其中GS组109例,CN-2组29例,分析两种表型在突变位点上的差异。计量资料两组间比较采用Mann-Whitney U检验。计数资料两组间比较采用χ2检验或Fisher精确检验。使用SNPStats软件对突变位点进行连锁不平衡(LD)分析及单倍型分析。强LD被定义为|D′|和r2均>0.8,中度LD被定义为|D′|>0.8且r2>0.4。 结果 138例患者均进行了UGT1A1基因检测,突变主要发生在启动子上游苯巴比妥反应增强元件(PBREM)区域-3279T>G突变(104例,75.36%)、-3152G>A突变(82例,59.42%)和启动子TATA盒TA插入突变(88例,63.77%),以及编码区1号外显子上的c.211 G>A突变(66例,47.83%)。GS组在PBREM区域-3279T>G位点(82.57% vs 48.28%,χ2=14.508,P<0.001)、-3152G>A位点(68.81% vs 24.14%,χ2=18.955,P<0.001)及启动子TATA盒(TA)6>(TA)7突变的比例(72.48% vs 31.03%,χ2=17.027,P<0.001)均明显高于CN-2组,而CN-2组在c.211位点(68.97% vs 42.20%,χ2=6.575,P=0.010)和c.1456位点(51.72% vs 7.34%,χ2=29.372,P<0.001)的突变比例显著高于GS组。对UGT1A1基因不同突变位点的LD分析发现,(TA)6>(TA)7与-3152G>A之间存在强LD(|D′|>0.8,r2>0.8),在(TA)6>(TA)7和-3279T>G、-3152G>A和-3279T>G、(TA)6>(TA)7和c.211G>A、-3279T>G和c.211G>A之间存在中度LD(|D′|均>0.8,r2均>0.4)。单倍型频率分析发现在GS组中单倍型-3279G—-3152A—(TA)7出现的频率高于CN-2组(45.72% vs 17.24%,χ2=7.833,P=0.005),而c.1456G(4.10% vs 16.48%,χ2=4.873,P=0.027)和c.211A—c.1456G(1.86% vs 24.90%,χ2=15.210,P<0.001)的频率低于CN-2组。双倍型分析发现单倍型c.1456G和c.211A—c.1456G参与组成的双倍型对应更高的TBil水平。 结论 GS和CN-2患者的常见UGT1A1基因突变位点以及主要单倍型不同,CN-2常见双倍型对应更高的TBil水平。 -
关键词:
- 吉尔伯特病 /
- Crigler-Najjar综合征 /
- 多态现象, 遗传
Abstract:Objective To investigate the differences in UGT1A1 gene mutation sites, haplotypes, and diplotypes between patients with Gilbert syndrome (GS) and those with Crigler-Najjar syndrome type Ⅱ (CN-2). Methods A retrospective analysis was performed for the clinical data of 138 patients with GS or CN-2 who attended Beijing YouAn Hospital, Capital Medical University, from January 1, 2010 to December 31, 2019, with 109 patients in the GS group and 29 patients in the CN-2 group, and the differences in mutation sites were analyzed between the two phenotypes. The Mann-Whitney U test was used for comparison of continuous data between two groups, and the chi-square test or the Fisher's exact test was used for comparison of categorical data between groups. SNPStats software was used to perform linkage disequilibrium (LD) and haplotype analyses of mutation sites. Strong LD was defined as both |D′| and r2 > 0.8, and moderate LD was defined as |D′| > 0.8 and r2 > 0.4. Results UGT1A1 gene detection was performed for all patients, and mutations mainly included -3279T > G mutation (104 patients, 75.36%) and -3152G > A mutation (82 patients, 59.42%) in the upstream promoter PBREM region, a promoter TATA box TA insertion mutation (88 patients, 63.77%), and c.211G > A mutation in Exon 1 of the coding region (66 patients, 47.83%). Compared with the CN-2 group, the GS group had a significantly higher proportion of PBREM region -3279T > G mutation (82.57% vs 48.28%, χ2=14.508, P < 0.001), PBREM region -3152G > A mutation (68.81% vs 24.14%, χ2=18.955, P < 0.001), and promoter TATA box (TA)6 > (TA)7 mutation (72.48% vs 31.03%, χ2=17.027, P < 0.001), and compared with the GS group, the CN-2 group had a significantly higher proportion of mutations at the c.211 locus (68.97% vs 42.20%, χ2=6.575, P=0.010) and the c.1456 locus (51.72% vs 7.34%, χ2=29.372, P < 0.001). LD analysis of different mutation sites of the UGT1A1 gene showed strong LD (|D′| > 0.8, r2 > 0.8) between (TA)6 > (TA)7 and -3152G > A and moderate LD (|D′| > 0.8, r2 > 0.4) between (TA)6 > (TA)7 and -3279T > G, between -3152G > A and -3279T > G, between (TA)6 > (TA)7 and c.211G > A, and between -3279T > G and c.211G > A. Haplotype frequency analysis showed that compared with the CN-2 group, the GS group had a significantly higher frequency of haplotype -3279G—-3152A—(TA)7 (45.72% vs 17.24%, χ2=7.833, P=0.005) and significantly lower frequencies of c.1456G (4.10% vs 16.48%, χ2=4.873, P=0.027) and c.211A—c.1456G (1.86% vs 24.90%, χ2=15.210, P < 0.001). The diplotype analysis showed that diplotypes consisting of haplotype c.1456G or c.211A—c.1456G were associated with a higher level of total bilirubin (TBil). Conclusion There are differences in common mutation sites and major haplotypes of the UGT1A1 gene between patients with GS and those with CN-2, and the common diplotypes of CN-2 correspond to a higher level of TBil. -
Key words:
- Gilbert Disease /
- Crigler-Najjar Syndrome /
- Polymorphism, Genetic
-
表 1 GS组与CN-2组一般资料比较
指标 GS组(n=109) CN-2组(n=29) 统计值 P值 男/女(例) 95/14 20/9 χ2=4.226 0.040 年龄(岁) 36.00(21.50~47.50) 29.00(19.00~36.00) Z=-1.971 0.049 ALT(U/L) 19.00(12.69~29.25) 22.00(11.35~35.55) Z=-0.920 0.358 AST(U/L) 20.00(17.00~25.00) 19.00(14.95~28.00) Z=-0.013 0.990 TBil(μmol/L) 46.70(34.55~62.80) 145.70(115.29~261.20) Z=-8.260 <0.001 IBil(μmol/L) 34.20(24.25~51.35) 133.10(101.30~226.30) Z=-8.082 <0.001 GGT(U/L) 18.00(13.00~25.00) 24.00(13.00~37.00) Z=1.020 0.308 ALP(U/L) 72.00(58.50~98.50) 91.00(66.00~124.00) Z=1.335 0.182 TP(g/L) 73.50(70.25~76.60) 69.50(62.60~74.05) Z=-3.217 0.001 Alb(g/L) 47.10(45.50~49.05) 45.70(40.63~49.10) Z=-2.271 0.023 表 2 UGT1A1基因各突变位点发生情况
突变位点 GS组
(n=109)CN-2组
(n=29)χ2值 P值 (TA)6>(TA)7[例(%)] 79(72.48) 9(31.03) 17.027 <0.001 -3279T>G[例(%)] 90(82.57) 14(48.28) 14.508 <0.001 -3152G>A[例(%)] 75(68.81) 7(24.14) 18.955 <0.001 c.211G>A[例(%)] 46(42.20) 20(68.97) 6.575 0.010 c.686C>A[例(%)] 11(10.09) 2(6.90) 0.028 0.868 c.1091C>T[例(%)] 5(4.59) 2(6.90) 0.001 0.978 c.1456T>G[例(%)] 8(7.34) 15(51.72) 29.372 <0.001 表 3 UGT1A1基因不同变异间的LD分析
项目 -3279T>G -3152G>A c.211G>A c.686C>A c.1091C>T c.1456T>G |D′|值 (TA)6>(TA)7 0.999 6 0.983 7 0.999 7 0.997 8 0.393 5 0.999 1 -3279T>G 0.999 6 0.978 9 0.997 3 0.995 6 0.999 3 -3152G>A 0.999 7 0.691 3 0.243 5 0.999 0 c.211G>A 0.996 5 0.994 2 0.347 7 c.686C>A 0.963 8 0.991 7 c.1091C>T 0.986 3 r2值 (TA)6>(TA)7 0.683 6 0.824 5 0.441 4 0.056 4 0.004 8 0.145 8 -3279T>G 0.582 3 0.618 6 0.038 5 0.023 8 0.213 3 -3152G>A 0.376 0 0.031 8 0.001 6 0.124 2 c.211G>A 0.024 8 0.015 4 0.040 0 c.686C>A 0.001 6 0.008 1 c.1091C>T 0.005 0 表 4 UGT1A1基因单倍型分布
单倍型 (TA)6
>(TA)7-3279T
>G-3152G
>Ac.211G
>Ac.686C
>Ac.1091C
>Tc.1456T
>G总数
(%)GS组
(%)CN-2组
(%)χ2值 P值 1 7 G A G C C T 39.70 45.72 17.24 7.833 0.005 2 6 T G A C C T 24.80 25.63 21.65 0.308 0.579 3 6 T G G C C G 6.76 4.10 16.48 4.873 0.027 4 6 T G A C C G 6.64 1.86 24.90 15.210 <0.001 5 6 G G G C C T 5.90 6.26 5.17 0.026 0.871 6 7 G A G A C T 3.96 4.48 1.72 0.379 0.538 7 7 G G G C C T 3.63 3.22 5.17 0.000 >0.05 8 6 T G G C C T 3.44 3.63 2.49 0.000 >0.05 9 6 G G G C T T 2.71 2.42 3.45 >0.05 -
[1] WATCHKO JF, TIRIBELLI C. Bilirubin-induced neurologic damage-mechanisms and management approaches[J]. N Engl J Med, 2013, 369(21): 2021-2030. DOI: 10.1056/NEJMra1308124. [2] DHAWAN A, LAWLOR MW, MAZARIEGOS GV, et al. Disease burden of Crigler-Najjar syndrome: Systematic review and future perspectives[J]. J Gastroenterol Hepatol, 2020, 35(4): 530-543. DOI: 10.1111/jgh.14853. [3] MARUO Y, NAKAHARA S, YANAGI T, et al. Genotype of UGT1A1 and phenotype correlation between Crigler-Najjar syndrome type Ⅱ and Gilbert syndrome[J]. J Gastroenterol Hepatol, 2016, 31(2): 403-408. DOI: 10.1111/jgh.13071. [4] BIANCO A, DVO RŘÁK A, CAPKOVÁ N, et al. The extent of intracellular accumulation of bilirubin determines its anti- or pro-oxidant effect[J]. Int J Mol Sci, 2020, 21(21): 8101. DOI: 10.3390/ijms21218101. [5] VÍTEK L. The role of bilirubin in diabetes, metabolic syndrome, and cardiovascular diseases[J]. Front Pharmacol, 2012, 3: 55. DOI: 10.3389/fphar.2012.00055. [6] LAPENNA D, CIOFANI G, PIERDOMENICO SD, et al. Association of serum bilirubin with oxidant damage of human atherosclerotic plaques and the severity of atherosclerosis[J]. Clin Exp Med, 2018, 18(1): 119-124. DOI: 10.1007/s10238-017-0470-5. [7] ZULUS B, GRVNBACHER G, KLEBER ME, et al. The UGT1A1*28 gene variant predicts long-term mortality in patients undergoing coronary angiography[J]. Clin Chem Lab Med, 2018, 56(4): 560-564. DOI: 10.1515/cclm-2017-0692. [8] AONO S, YAMADA Y, KEINO H, et al. Identification of defect in the genes for bilirubin UDP-glucuronosyl-transferase in a patient with Crigler-Najjar syndrome type Ⅱ[J]. Biochem Biophys Res Commun, 1993, 197(3): 1239-1244. DOI: 10.1006/bbrc.1993.2610. [9] ARIAS IM, LONDON IM. Bilirubin glucuronide formation in vitro; demonstration of a defect in Gilbert's disease[J]. Science, 1957, 126(3273): 563-564. DOI: 10.1126/science.126.3273.563. [10] BOSMA PJ, CHOWDHURY JR, BAKKER C, et al. The genetic basis of the reduced expression of bilirubin UDP-glucurono-syltransferase 1 in Gilbert's syndrome[J]. N Engl J Med, 1995, 333(18): 1171-1175. DOI: 10.1056/NEJM199511023331802. [11] VITEK L, BELLAROSA C, TIRIBELLI C. Induction of mild hyperbilirubinemia: Hype or real therapeutic opportunity?[J]. Clin Pharmacol Ther, 2019, 106(3): 568-575. DOI: 10.1002/cpt.1341. -




 PDF下载 ( 2018 KB)
PDF下载 ( 2018 KB)

 下载:
下载:


