华东沿海地区多中心40例胰腺肿瘤患者基因组图谱分析
DOI: 10.3969/j.issn.1001-5256.2022.02.028
利益冲突声明:本研究不存在研究者、伦理委员会成员、受试者监护人以及与公开研究成果有关的利益冲突。
作者贡献声明:王晶、谭斌负责课题设计,资料分析,撰写论文;赵志杰、仲灏辰参与收集数据,修改论文;曲林林负责拟定写作思路,指导撰写文章并最后定稿。
Genomic profile of pancreatic tumor in the coastal regions of Eastern China: A multicenter analysis of 40 cases
-
摘要:
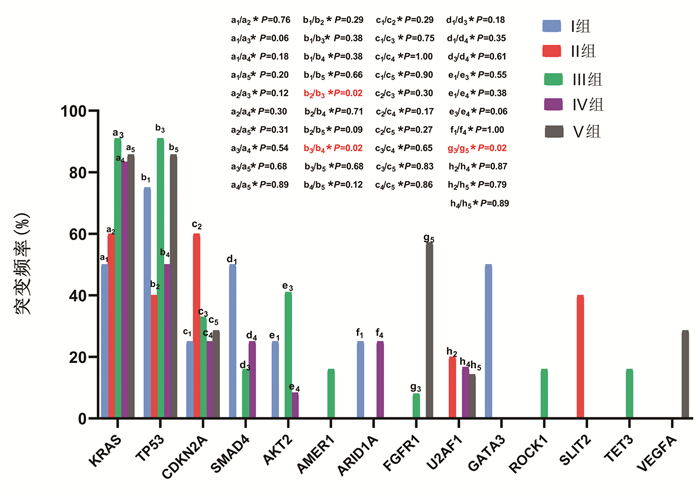
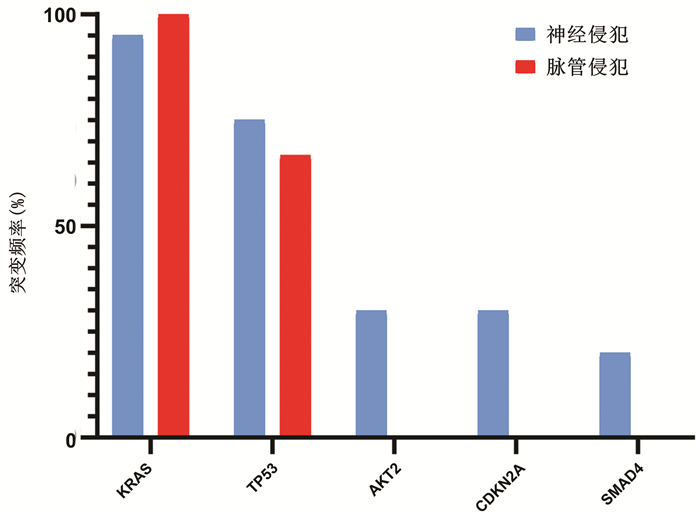
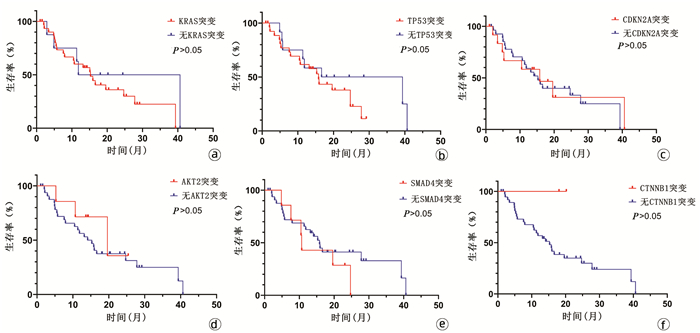
目的 探讨我国华东沿海地区胰腺癌患者的基因突变情况,为个体化治疗提供依据。 方法 选取2017年1月—2019年6月在青岛大学附属医院、青岛市立医院、烟台山医院、烟台中法友好医院收治并经手术治疗后诊断为胰腺恶性肿瘤的患者40例。采用下一代测序技术检测肿瘤组织和体细胞中的基因突变。绘制基因突变图谱,分析基因组改变。计数资料组间比较采用χ2检验或Fisher精确检验。采用Kaplan-Meier法绘制生存曲线,组间比较采用log-rank检验。 结果 40例患者中胰腺导管腺癌34例(85.0%),胰腺实性假乳头状肿瘤3例(7.5%),胰腺神经内分泌肿瘤1例(2.5%),分型不清2例(5.0%)。KRAS(80.0%,32/40)、TP53(70.0%,28/40)、CDKN2A(32.5%,13/40)、SMAD4(17.5%,7/40)和AKT2(17.5%,7/40)是最常见的突变。5种常见基因突变的生存时间差异均无统计学意义(P值均>0.05)。 结论 下一代测序技术能够提供全面、准确的基因组改变信息,可为胰腺癌的诊断和精确治疗提供新的潜在生物标志物。通过对突变基因进行分析,从而为胰腺癌的个体化治疗奠定基础。 Abstract:Objective To investigate the gene mutations of Chinese patients with pancreatic cancer in the coastal regions of Eastern China, and to provide a basis for individualized treatment. Methods A total of 40 patients who were admitted and diagnosed with malignant pancreatic tumor after surgical treatment in The Affiliated Hospital of Qingdao University, Qingdao Municipal Hospital, Yantaishan Hospital, and Yantai Sino-France Friendship Hospital from January 2017 to June 2019 were enrolled. Next-generation sequencing (NGS) was used to detect gene mutations in tumor tissue and somatic cells, and the map of gene mutations was plotted to analyze genomic alterations. The chi-square test or the Fisher's exact test was used for comparison of categorical data between groups. The Kaplan-Meier method was used to plot survival curves, and the log-rank test was used for comparison between groups. Results Among the 40 patients, 34 (85.0%) had pancreatic ductal adenocarcinoma, 3 (7.5%) had solid pseudopapillary neoplasm of the pancreas, 1 (2.5%) had pancreatic neuroendocrine tumor, and 2 (5.0%) had unclear typing. KRAS (80.0%, 32/40), TP53 (70.0%, 28/40), CDKN2A (32.5%, 13/40), SMAD4 (17.5%, 7/40), and AKT2 (17.5%, 7/40) were the most common mutations, and there was no significant difference in survival time between the patients with these five common gene mutations (all P > 0.05). Conclusion NGS technology can provide comprehensive and accurate information of genomic alterations and may provide novel potential biomarkers for the diagnosis and precise treatment of pancreatic cancer. The analysis of mutant genes also lays a foundation for the individualized treatment of pancreatic cancer. -
Key words:
- Pancreatic Neoplasms /
- Genome /
- Mutation
-
[1] SIEGEL RL, MILLER KD, JEMAL A. Cancer statistics, 2019[J]. CA Cancer J Clin, 2019, 69(1): 7-34. DOI: 10.3322/caac.21551. [2] STEUER CE, RAMALINGAM SS. Tumor mutation burden: Leading immunotherapy to the era of precision medicine?[J]. J Clin Oncol, 2018, 36(7): 631-632. DOI: 10.1200/JCO.2017.76.8770. [3] von HOFF DD, ERVIN T, ARENA FP, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine[J]. N Engl J Med, 2013, 369(18): 1691-1703. DOI: 10.1056/NEJMoa1304369. [4] CONROY T, PAILLOT B, FRANÇOIS E, et al. Irinotecan plus oxaliplatin and leucovorin-modulated fluorouracil in advanced pancreatic cancer—a Groupe Tumeurs Digestives of the Federation Nationale des Centres de Lutte Contre le Cancer study[J]. J Clin Oncol, 2005, 23(6): 1228-1236. DOI: 10.1200/JCO.2005.06.050. [5] FRANCO J, WITKIEWICZ AK, KNUDSEN ES. CDK4/6 inhibitors have potent activity in combination with pathway selective therapeutic agents in models of pancreatic cancer[J]. Oncotarget, 2014, 5(15): 6512-6525. DOI: 10.18632/oncotarget.2270. [6] OSHIMA M, OKANO K, MURAKI S, et al. Immunohistochemically detected expression of 3 major genes (CDKN2A/p16, TP53, and SMAD4/DPC4) strongly predicts survival in patients with resectable pancreatic cancer[J]. Ann Surg, 2013, 258(2): 336-346. DOI: 10.1097/SLA.0b013e3182827a65. [7] BIANKIN AV, WADDELL N, KASSAHN KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes[J]. Nature, 2012, 491(7424): 399-405. DOI: 10.1038/nature11547. [8] KAWAGUCHI K, IGARASHI K, MIYAKE K, et al. MEK inhibitor trametinib in combination with gemcitabine regresses a patient-derived orthotopic xenograft (PDOX) pancreatic cancer nude mouse model[J]. Tissue Cell, 2018, 52: 124-128. DOI: 10.1016/j.tice.2018.05.003. [9] BOURNET B, BUSCAIL C, MUSCARI F, et al. Targeting KRAS for diagnosis, prognosis, and treatment of pancreatic cancer: Hopes and realities[J]. Eur J Cancer, 2016, 54: 75-83. DOI: 10.1016/j.ejca.2015.11.012. [10] MORTON JP, TIMPSON P, KARIM SA, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer[J]. Proc Natl Acad Sci U S A, 2010, 107(1): 246-251. DOI: 10.1073/pnas.0908428107. [11] Cancer Genome Atlas Research Network. Integrated genomic characterization of pancreatic ductal adenocarcinoma[J]. Cancer Cell, 2017, 32(2): 185-203. DOI: 10.1016/j.ccell.2017.07.007. [12] WADDELL N, PAJIC M, PATCH AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer[J]. Nature, 2015, 518(7540): 495-501. DOI: 10.1038/nature14169. [13] ORMANNS S, HAAS M, REMOLD A, et al. The impact of SMAD4 loss on outcome in patients with advanced pancreatic cancer treated with systemic chemotherapy[J]. Int J Mol Sci, 2017, 18(5): 1094. DOI: 10.3390/ijms18051094. [14] XIA X, WU W, HUANG C, et al. SMAD4 and its role in pancreatic cancer[J]. Tumour Biol, 2015, 36(1): 111-119. DOI: 10.1007/s13277-014-2883-z. [15] CRANE CH, VARADHACHARY GR, YORDY JS, et al. Phase Ⅱ trial of cetuximab, gemcitabine, and oxaliplatin followed by chemoradiation with cetuximab for locally advanced (T4) pancreatic adenocarcinoma: Correlation of Smad4(Dpc4) immunostaining with pattern of disease progression[J]. J Clin Oncol, 2011, 29(22): 3037-3043. DOI: 10.1200/JCO.2010.33.8038. [16] OTTENHOF NA, MORSINK FH, TEN KATE F, et al. Multivariate analysis of immunohistochemical evaluation of protein expression in pancreatic ductal adenocarcinoma reveals prognostic significance for persistent Smad4 expression only[J]. Cell Oncol (Dordr), 2012, 35(2): 119-126. DOI: 10.1007/s13402-012-0072-x. [17] SHIN SH, KIM HJ, HWANG DW, et al. The DPC4/SMAD4 genetic status determines recurrence patterns and treatment outcomes in resected pancreatic ductal adenocarcinoma: A prospective cohort study[J]. Oncotarget, 2017, 8(11): 17945-17959. DOI: 10.18632/oncotarget.14901. [18] CHENG JQ, RUGGERI B, KLEIN WM, et al. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA[J]. Proc Natl Acad Sci U S A, 1996, 93(8): 3636-3641. DOI: 10.1073/pnas.93.8.3636. [19] RUGGERI BA, HUANG L, WOOD M, et al. Amplification and overexpression of the AKT2 oncogene in a subset of human pancreatic ductal adenocarcinomas[J]. Mol Carcinog, 1998, 21(2): 81-86. [20] CUI Y, WANG Q, WANG J, et al. Knockdown of AKT2 expression by RNA interference inhibits proliferation, enhances apoptosis, and increases chemosensitivity to the anticancer drug VM-26 in U87 glioma cells[J]. Brain Res, 2012, 1469: 1-9. DOI: 10.1016/j.brainres.2012.06.043. [21] NITULESCU GM, MARGINA D, JUZENAS P, et al. Akt inhibitors in cancer treatment: The long journey from drug discovery to clinical use (Review)[J]. Int J Oncol, 2016, 48(3): 869-885. DOI: 10.3892/ijo.2015.3306. [22] CHEN D, NIU M, JIAO X, et al. Inhibition of AKT2 enhances sensitivity to gemcitabine via regulating PUMA and NF-κB signaling pathway in human pancreatic ductal adenocarcinoma[J]. Int J Mol Sci, 2012, 13(1): 1186-1208. DOI: 10.3390/ijms13011186. [23] LIU Q, TURNER KM, ALFRED YUNG WK, et al. Role of AKT signaling in DNA repair and clinical response to cancer therapy[J]. Neuro Oncol, 2014, 16(10): 1313-1323. DOI: 10.1093/neuonc/nou058. [24] KORINEK V, BARKER N, MORIN PJ, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma[J]. Science, 1997, 275(5307): 1784-1787. DOI: 10.1126/science.275.5307.1784. [25] LOGAN CY, NUSSE R. The Wnt signaling pathway in development and disease[J]. Annu Rev Cell Dev Biol, 2004, 20: 781-810. DOI: 10.1146/annurev.cellbio.20.010403.113126. [26] CLEVERS H. Wnt/beta-catenin signaling in development and disease[J]. Cell, 2006, 127(3): 469-480. DOI: 10.1016/j.cell.2006.10.018. [27] SINGHI AD, GEORGE B, GREENBOWE JR, et al. Real-time targeted genome profile analysis of pancreatic ductal adenocarcinomas identifies genetic alterations that might be targeted with existing drugs or used as biomarkers[J]. Gastroenterology, 2019, 156(8): 2242-2253. DOI: 10.1053/j.gastro.2019.02.037. [28] WEISS GJ, BLAYDORN L, BECK J, et al. Phase Ib/Ⅱ study of gemcitabine, nab-paclitaxel, and pembrolizumab in metastatic pancreatic adenocarcinoma[J]. Invest New Drugs, 2018, 36(1): 96-102. DOI: 10.1007/s10637-017-0525-1. 期刊类型引用(1)
1. 王淼东,彭佩纯,陈泽山,邓鑫. 基于生物信息学研究肝癌脂肪酸代谢lncRNA预后免疫反应及药物敏感性分析. 中国免疫学杂志. 2024(12): 2565-2573 .  百度学术
百度学术其他类型引用(0)
-
 王晶-华东沿海地区多中心40例胰腺肿瘤患者基因组图谱分析-附录1.pdf
王晶-华东沿海地区多中心40例胰腺肿瘤患者基因组图谱分析-附录1.pdf

-




 PDF下载 ( 3577 KB)
PDF下载 ( 3577 KB)


 下载:
下载:
 百度学术
百度学术

