长链非编码RNA LNC01309对人肝癌细胞增殖和迁移能力的影响及作用机制
DOI: 10.3969/j.issn.1001-5256.2022.03.014
Effect of long non-coding RNA LNC 01309 on proliferation and migration abilities of human hepatoma cells and its mechanism of action
-
摘要:
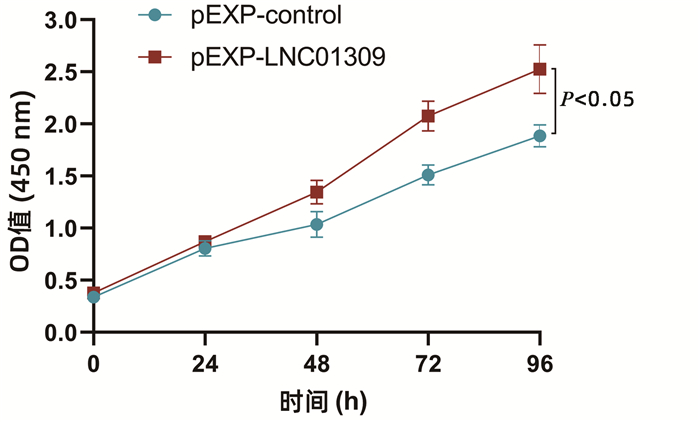
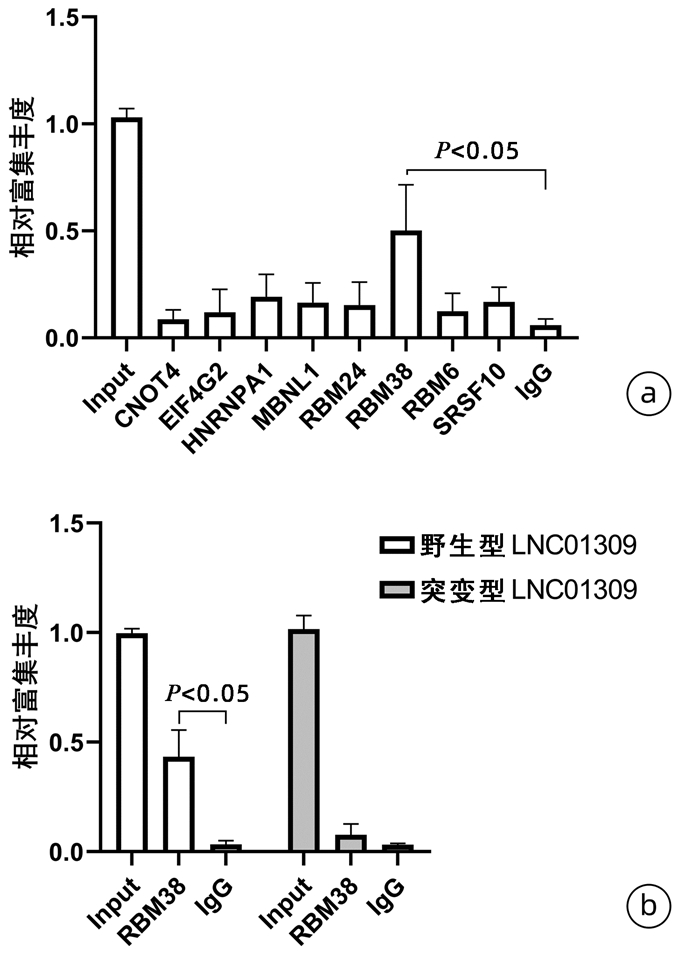
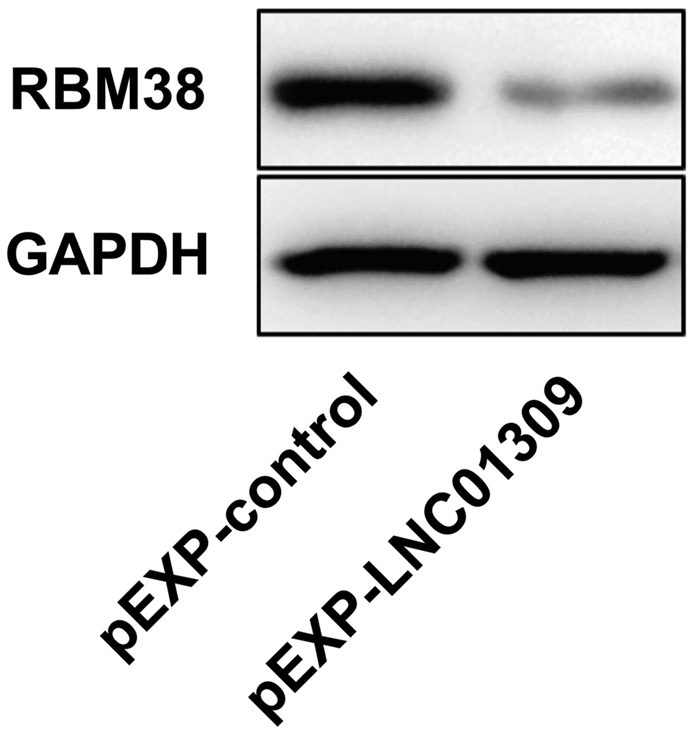
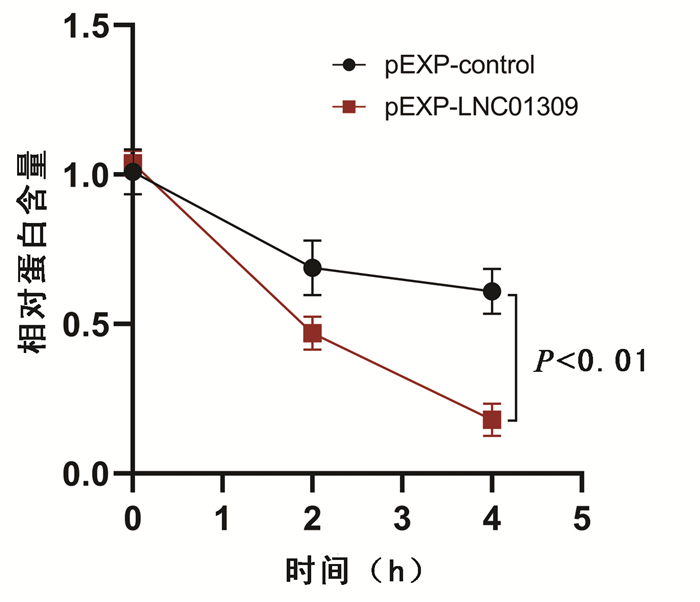
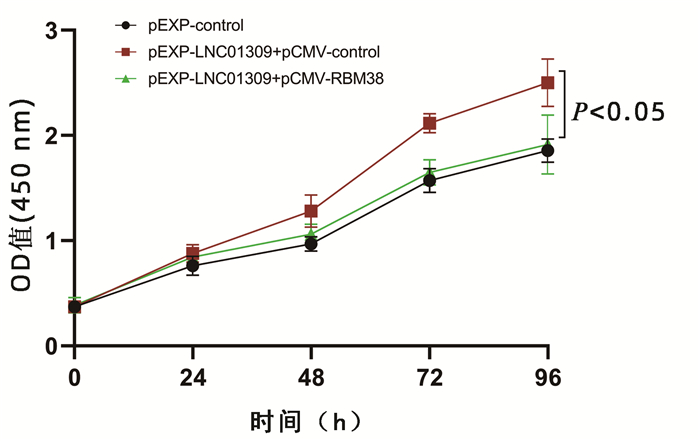
目的 探讨长链非编码RNA(lncRNA)LNC01309对人肝癌细胞增殖和迁移能力的影响及作用机制。 方法 收集2018年2月—2019年6月在解放军总医院第六医学中心手术治疗的12例肝细胞癌(以下简称肝癌)患者标本及对应的癌旁组织,采用荧光定量PCR方法检测LNC01309的相对表达量。利用荧光定量PCR法检测LNC01309在人肝癌细胞系(Hep G2、SNU-398和Hep 3B)和人永生化正常肝细胞系(THLE-2)中的表达水平。在肝癌细胞Hep G2中过表达LNC01309,分为质粒对照组(pEXP-control)和过表达组(pEXP-LNC01309)。采用CCK-8检测各组细胞增殖变化,采用划痕愈合试验和Transwell试验检测各组细胞迁移能力。利用RNA免疫共沉淀试验检测LNC01309与RBM38的相互作用(分组为IgG组和RBM38抗体组)。利用放线菌酮追踪分析检测LNC01309对于RBM38蛋白稳定性的影响。在肝癌细胞Hep G2中过表达RBM38进行回复试验,采用CCK-8试验、划痕愈合试验和Transwell试验,分别检测细胞增殖和迁移能力的变化。计量资料两组间比较采用t检验。 结果 LNC01309在肝癌组织中的平均表达水平高于癌旁组织(4.225±2.285 vs 1.541±0.530, t=3.618,P=0.004)。LNC01309在肝癌细胞(Hep G2、SNU-398和Hep 3B)中的相对表达水平亦均高于正常肝细胞(THLE-2)(t值分别为4.231、6.489、14.480,P值均<0.05)。过表达LNC01309的Hep G2细胞(t=9.172,P<0.001)相对于对照组,细胞的生长速度明显上升(第96小时OD450值:1.885±0.107 vs 2.527±0.234,t=4.330,P=0.012),迁移能力上调(划痕愈合试验:11.65%±2.40% vs 35.66%±4.90%,t=9.837,P<0.001;Transwell试验:100.00%±3.11% vs 161.00%±35.93%,t=4.399,P=0.005);然而过表达LNC01309诱导的肝癌细胞增殖和迁移能力的上调效应,都能够被RBM38部分抑制(第96小时OD450值:2.500±0.227 vs 1.913±0.282,t=2.812,P=0.048;168.00%±9.43% vs 117.20%±18.03%,t=6.622,P<0.001)。相对于IgG对照组,RBM38抗体能够显著富集沉淀LNC01309(t=3.846,P=0.031)。放线菌酮试验结果显示,过表达LNC01309组细胞中RBM38蛋白稳定性显著下调(t=8.038,P=0.001)。 结论 新发现的LNC01309通过与RBM38的相互作用降低了RBM38蛋白稳定性,促进肝癌细胞的增殖与迁移。 -
关键词:
- 癌, 肝细胞 /
- RNA, 长链非编码 /
- RNA结合蛋白质类
Abstract:Objective To investigate the effect of long non-coding RNA (lncRNA) LNC01309 on the proliferation and migration abilities of human hepatocellular carcinoma (HCC) cells and its mechanism of action. Methods HCC samples and corresponding adjacent tissue samples were collected from 12 patients with HCC who underwent surgical treatment in The Sixth Medical Center of PLA General Hospital from February 2018 to June 2019, and quantitative real-time PCR was used to measure the relative expression level of LNC01309. Quantitative real-time PCR was also used to measure the expression level of LNC01309 in human hepatoma cell lines (HepG2, SNU-398, and Hep3B) and the human immortalized normal liver cell line THLE-2. After LNC01309 was overexpressed in HepG2 cells, the cells were divided into plasmid control group (pEXP-control) and overexpression group (pEXP-LNC01309). CCK-8 assay was used to observe the change in cell proliferation, and wound healing assay and Transwell assay were used to observe migration ability. RNA co-immunoprecipitation was used to detect the interaction between LNC01309 with RBM38, with cells divided into IgG group and RBM38 antibody group, and cycloheximide chase assay was used to analyze the effect of LNC01309 on the stability of RBM38 protein. RBM38 was overexpressed in HepG2 cells to conduct the recovery experiment, and CCK-8 assay, wound healing assay, and Transwell assay were used to observe the changes in cell proliferation and migration abilities. The t-test was used for comparison of continuous data between two groups. Results The mean expression level of LNC01309 in HCC tissue was significantly higher than that in adjacent tissue (4.225±2.285 vs 1.541±0.530, t=3.618, P=0.004), and the relative expression level of LNC01309 in hepatoma cells (HepG2, SNU-398, and Hep3B) was also significantly higher than that in normal hepatocytes (THLE-2) (t=4.231、6.489、14.480, all P < 0.05). Compared with the control group, HepG2 cells with the overexpression of LNC01309 had significant increases in growth rate (OD450 value at 96 hours: 1.885±0.107 vs 2.527±0.234, t=4.330, P=0.012) and migration ability (11.65%±2.40% vs 35.66%±4.90%, t=9.837, P < 0.001; 100.00%±3.11% vs 161.00%±35.93%, t=4.399, P=0.005); however, the upregulated proliferation and migration abilities of hepatoma cells induced by LNC01309 overexpression were partially inhibited by RBM38 (OD450 value at 96 hours: 2.500±0.227 vs 1.913±0.282, t=2.812, P=0.048; 168.00%±9.43% vs 117.20%±18.03%, t=6.622, P < 0.001). Compared with the IgG control group, RBM38 antibody significantly enriched the precipitation of LNC01309 (t=3.846, P=0.031). The results of cycloheximide chase assay showed that the LNC01309 overexpression group had a significant reduction in the stability of RBM38 protein (t=8.038, P=0.001). Conclusion The newly identified LNC01309 reduces the stability of RBM38 protein through interaction with RBM38 and promotes the proliferation and migration of HCC cells. -
Key words:
- Carcinoma, Hepatocellular /
- RNA, Long Noncoding /
- RNA-Binding Proteins
-
肝细胞癌(HCC)是我国最常见的恶性肿瘤之一。国家癌症中心最新数据显示,我国每年新发肝癌约37万人,约32.6万人死于肝癌,其中94.7%与慢性HBV感染有关[1-2]。早期发现、早期诊断和早期治疗是改善原发性肝癌预后的关键。然而目前尚缺乏原发性肝癌高敏感度和高特异度的诊断指标。自噬是细胞自我吞噬胞内长寿蛋白或受损细胞器并通过溶酶体途径进行的分解代谢过程,从而维持细胞稳态[3]。近年研究[4-7]显示自噬在HBV感染和HCC的发生发展起重要作用。但目前关于自噬相关蛋白在HBV相关HCC(HBV-HCC)患者血清中的表达及意义鲜有报道。本研究通过分析HBV-HCC、非HBV相关HCC(nonHBV-HCC)、慢性乙型肝炎(CHB)和健康对照者血清自噬相关蛋白7(ATG7)的表达水平,探讨ATG7对HBV-HCC的诊断价值。
1. 资料与方法
1.1 研究对象
选取2018年6月—2020年12月于本院住院的HCC患者89例。HCC的诊断标准符合《原发性肝癌诊疗规范(2017年版)》[8]并经病理确认。排除标准如下:病理类型为肝内胆管癌或HCC-肝内胆管癌混合型者;已接受过肝移植术、局部消融、肝动脉化疗栓塞术、放化疗等抗肿瘤治疗者;继发性HCC患者;同时伴有其他肿瘤患者。CHB诊断符合《慢性乙型肝炎防治指南(2019年版)》[9]。根据血清是否检出HBsAg和/或HBV DNA分为HBV-HCC组及nonHBV-HCC组。选取同期50例CHB患者(CHB组)和20例健康体检者作为对照组(HC组)。
1.2 资料收集
收集研究对象性别和年龄,总蛋白(TP)、白蛋白(Alb)、TBil、DBil、ALT、AST、GGT和ALP等实验室指标。另收集HBV-HCC组患者的BCLC分期和肿瘤直径等病理资料。
1.3 血清ATG7检测
所有研究对象清晨空腹肘静脉采血,采血后1 h内送实验室,3000 r/min离心10 min分离血清,-80 ℃冰箱保存。ATG7检测前4 h室温下复溶。使用上海江莱生物技术有限公司人ATG7酶联免疫吸附测定试剂盒(批号:Aug 2020)进行检测,严格按试剂盒说明书操作,在加入终止液15 min内用PHOMO酶标仪(安图生物,合肥)在450 nm波长测定各孔光密度(OD值),用WPS表格以标准品的浓度为纵坐标,OD值为横坐标绘制标准曲线并取得多项式公式,根据多项式公式计算每个样本的ATG7浓度。
1.4 伦理学审查
本研究方案经由福建医科大学孟超肝胆医院伦理委员会审批,批号:科审2020-029-01。
1.5 统计学方法
采用SPSS 22.0进行统计分析。非正态分布的计量资料以M(P25~P75)表示,多组间比较采用Kruskal-Wallis H检验,2组间比较采用Mann-Whitney U检验;计数资料组间比较采用χ2检验;采用Spearman进行相关性分析。运用GraphPad Prism7.0绘制散点图,采用Medcalc18.11.3绘制受试者工作特征曲线(ROC曲线)并比较不同指标曲线下面积(AUC)。P<0.05为差异有统计学意义。
2. 结果
2.1 一般资料
HCC、CHB及HC 3组比较,男女比例差异无统计学意义,其余指标差异均有统计学意义(P值均<0.01)(表 1)。89例HCC患者中,67例(75.28%)血清HBsAg和/或HBV DNA阳性,纳入HBV-HCC组;剩余22例(24.72%)纳入nonHBV-HCC组,包括5例酒精性肝硬化,8例非酒精性脂肪肝,1例慢性丙型肝炎,2例肝硬化(病因未明),6例慢性肝炎(病因未明)。HBV-HCC组血清TBil水平及具有肝硬化背景病例数高于nonHBV-HCC组,而nonHBV-HCC组的血清AST水平和最大肿瘤直径高于HBV-HCC组,差异均有统计学意义(P值均<0.05),其余参数差异均无统计学意义(P值均>0.05)(表 2)。
表 1 3组人口学和实验室特征比较参数 HCC组(n=89) CHB组(n=50) HC组(n=20) χ2值 P值 年龄(岁) 56.0(48.0~66.5) 42.0(32.8~52.5) 37.5(30.0~48.8) 38.807 <0.001 男性[例(%)] 77(86.52) 38(76.00) 13(65.00) 5.480 0.076 TBil(μmol/L) 19.30(12.65~26.10) 19.55(12.78~29.28) 12.15(9.48~18.63) 10.478 0.005 DBil(μmol/L) 7.60(4.70~10.85) 4.45(2.20~7.05) 5.30(4.15~6.35) 17.168 <0.001 TP(g/L) 61.0(54.5~67.5) 66.5(60.8~73.3) 72.0(70.0~77.0) 34.168 <0.001 Alb(g/L) 34.0(31.0~38.5) 37.5(34.0~41.0) 44.0(41.0~47.0) 42.516 <0.001 ALT(U/L) 115.0(62.0~218.5) 87.5(26.5~206.8) 19.0(12.3~26.5) 37.212 <0.001 AST(U/L) 149.0(54.0~286.0) 57.0(31.5~110.5) 17.0(14.3~20.0) 54.345 <0.001 GGT(U/L) 52.0(34.0~110.5) 68.5(29.3~112.0) 17.0(14.3~24.5) 31.527 <0.001 ALP(U/L) 81.0(60.5~103.0) 102.0(82.5~117.0) 66.5(52.8~88.3) 21.160 <0.001 AFP(ng/mL) 37.90(5.00~712.32) 7.20(3.05~241.27) 2.85(2.18~4.48) 33.048 <0.001 ATG7(ng/mL) 21.11(17.76~22.73) 19.21(16.65~20.82) 13.82(8.70~17.82) 33.134 <0.001 表 2 HBV-HCC组与nonHBV-HCC组生化和病理特征比较参数 HBV-HCC组(n=67) nonHBV-HCC组(n=22) 统计值 P值 年龄(岁) 55.0(46.0~ 65.0) 63.5(50.8~71.8) U=535.000 0.055 男/女(例) 58/9 19/3 χ2=0.001 0.981 TBil(μmol/L) 20.60(13.30~27.40) 16.00(11.15~21.48) U=524.500 0.043 DBil(μmol/L) 7.70(4.80~10.90) 7.15(3.80~9.65) U=602.500 0.201 TP(g/L) 61.0(50.0~67.0) 58.5(51.3~68.3) U=638.500 0.348 Alb(g/L) 34.0(31.0~38.0) 33.0(30.3~39.0) U=709.500 0.793 ALT(U/L) 95.0(54.0~198.0) 152.0(89.8~268.0) U=557.000 0.087 AST(U/L) 136.0(37.0~240.0) 236.5(93.8~348.8) U=523.000 0.042 GGT(U/L) 51.0(30.0~111.0) 58.0(35.0~113.3) U=658.000 0.452 ALP(U/L) 76.0(60.0~99.0) 92.0(62.8~127.8) U=575.500 0.124 AFP(ng/mL) 58.00(7.10~945.80) 18.28(2.48~405.33) U=536.500 0.057 BCLC(A/B/C/D,例) 51/4/12/0 12/5/5/0 χ2=5.229 0.073 最大肿瘤直径(cm) 3.95(2.50~7.70) 8.00(3.70~11.00) U=520.000 0.047 肝硬化[例(%)] 55(82.09) 9(40.91) χ2=13.904 <0.001 2.2 HBV-HCC血清ATG7表达水平升高
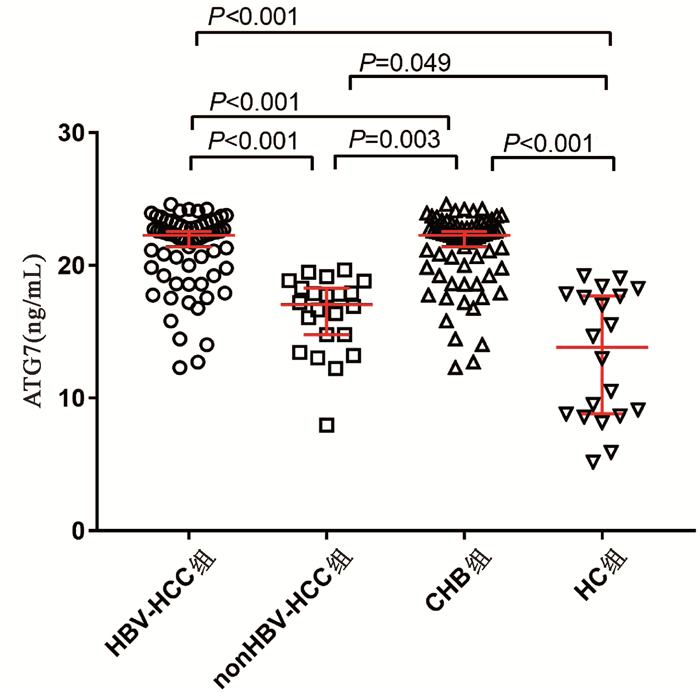
HBV- HCC组、nonHBV-HCC组、CHB组和HC组血清ATG7水平分别为22.88(19.79~23.04)ng/mL、17.06(14.45~19.40)ng/mL、19.21(16.65~20.82)ng/mL和13.82(8.70~17.82)ng/mL,差异有统计学意义(χ2=65.144,P<0.001)。两两比较显示HBV-HCC组血清ATG7不仅高于CHB组(U=758.5,P<0.001)和HC组(U=94.0,P<0.001),也高于nonHBV-HCC组(U=142.0,P<0.001);而nonHBV-HCC组血清ATG7水平不仅低于HBV-HCC组,也低于CHB组(U=311.0,P=0.003),仅稍高于HC组(U=142.0,P=0.049)(图 1)。此外,在67例HBV-HCC患者中,有肝硬化背景和无肝硬化背景患者的血清ATG7水平分别为22.32 (19.79~22.99)ng/mL和20.89(19.40~23.37)ng/mL,差异无统计学意义(P>0.05)。
2.3 血清ATG7表达水平和血清AFP水平的相关性
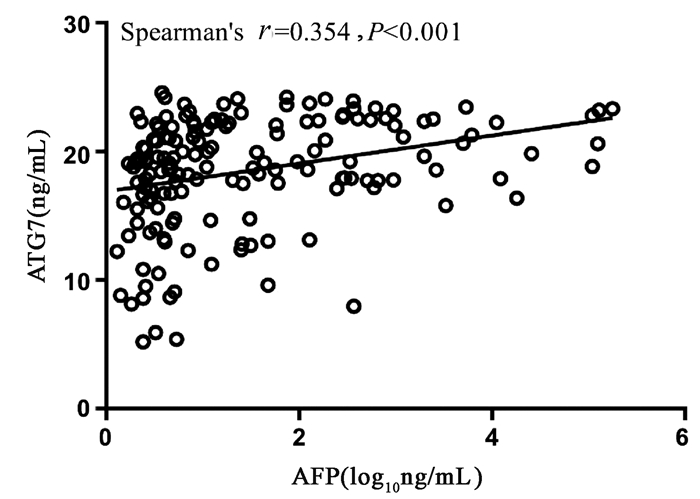
Spearman相关性分析显示,血清ATG7表达水平和血清AFP水平呈正相关,但相关性较弱(r=0.354,95%CI:0.205~0.486,P<0.001)(图 2)。
2.4 ATG7在HBV-HCC中的诊断性能
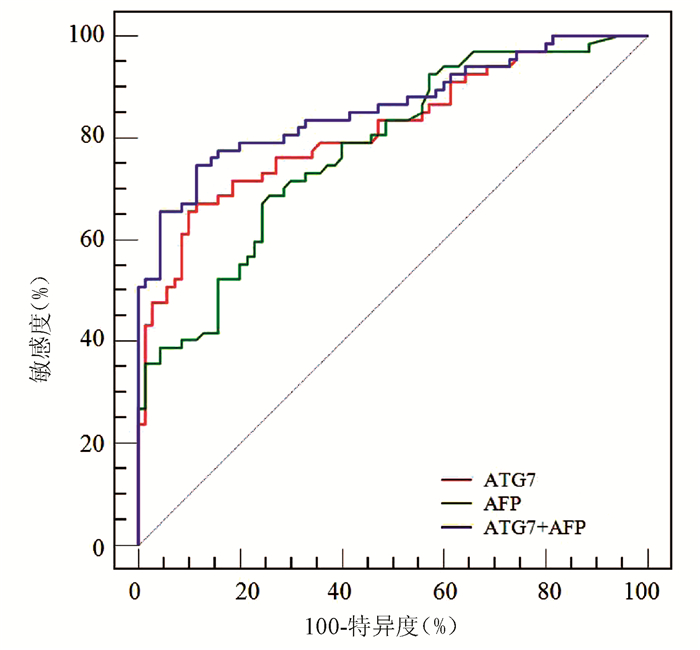
ROC曲线分析显示,ATG7诊断HBV-HCC的AUC为0.818(95% CI:0.743~0.879),稍高于AFP(AUC=0.777,95%CI:0.698~0.843),但二者差异无统计学意义(Z=0.852,P=0.394)(图 3);ATG7诊断HBV-HCC的cut-off值为20.08 ng/mL,敏感度和特异度分别为71.64%和77.14%,均略高于AFP(68.66%和74.29%)。ATG7联合AFP的二元logistic回归预测概率的AUC为0.859 (95%CI:0.790~0.913),显著高于ATG7(Z=2.192,P=0.028)和AFP(Z=2.076,P=0.038)(图 3, 表 3)。
表 3 ATG7、AFP及其联合检测诊断HBV-HCC的性能项目 AUC cut-off值 敏感度(%) 特异度(%) 阳性预测值(%) 阴性预测值(%) AFP 0.777 12.20 ng/mL 68.66 74.29 71.9 71.2 ATG7 0.818 20.08 ng/mL 71.64 77.14 75.0 74.0 ATG7+AFP 0.859 0.56 74.63 88.57 86.2 78.5 3. 讨论
本研究通过ELISA技术检测HCC患者血清ATG7表达水平,结果显示HBV-HCC患者血清ATG7显著升高,血清ATG7是诊断HBV-HCC的良好标志物,ATG7与AFP联合诊断可显著提高HBV-HCC的诊断效率。
ATG7作为一种泛素样修饰物激活酶,通过激活ATG8和ATG12调节自噬体的延伸,是自噬体形成过程的关键蛋白之一[10]。近年研究[11-12]提示ATG7及自噬在HBV-HCC发生发展中发挥重要作用。首先,HBV感染或HBV病毒蛋白的表达可诱导ATG7表达及自噬反应。本研究中CHB组血清ATG7显著高于健康对照组,印证了自噬与HBV感染有关。其次,多项研究[13-14]显示包括ATG7在内的多种自噬相关蛋白在HCC组织表达显著提高。近年的分子细胞学试验进一步揭示多种分子可通过ATG7调节自噬促进HCC发生发展[15-17]。研究[15]显示长链非编码RNA HOX转录反义RNA(HOTAIR)可通过上调ATG7表达激活自噬促进HCC细胞系增殖。癌性锚蛋白重复序列可直接与胞质中的ATG7蛋白相互作用诱导自噬从而促进HCC细胞进展[16]。肿瘤坏死因子α诱导蛋白8(TNFAIP8)则通过与ATG3-ATG7复合物相互作用调节自噬促进HCC细胞增殖[17]。相反,靶向敲减ATG7可抑制自噬,从而促进细胞凋亡、延缓细胞周期及抑制HCC细胞增殖[13]。上述研究结果表明ATG7与HBV-HCC密切相关。本研究数据显示HBV-HCC患者血清ATG7显著高于CHB和健康对照组,进一步证实了自噬与HBV-HCC的相关性,提示血清ATG7是一种潜在HBV-HCC标志物。而且HBV-HCC患者血清ATG7也显著高于nonHBV-HCC,提示不同病因的HCC发病机制不同,其血清标志物也可不同。
虽然AFP已在临床应用数十年,但诊断敏感度不高。最近的一项系统综述[18]显示,AFP诊断HCC的合并敏感度仅为63.9%。与此相符,本研究中AFP诊断HBV-HCC的敏感度、特异度和AUC分别为68.66%、74.29%和0.777。虽然ATG7的敏感度与AFP相仿,但其特异度较高,总体诊断性能较好。此外,ATG7和AFP相关性较弱,提示二者联合检测可有效提高诊断性能。早前有学者[19]提出应用logistic回归分析有助于提高多指标联合诊断效率。本研究结果显示ATG7联合AFP的二元logistic回归预测模型对HBV-HCC的整体诊断性能较好,AUC及诊断敏感度和特异度均显著高于ATG7及AFP。
总之,本研究显示ATG7是HBV-HCC的良好标志物,与AFP有较好的互补性,二者联合检测可显著提高HBV-HCC的诊断效率。然而本研究为单中心横断面研究,纳入研究病例有限,因此其诊断性能仍有待进一步扩大样本的多中心临床验证。
-
注:a,LNC01309与lncRNA MIR205HG序列相似性,并根据序列差异性设计特异性检测引物;b,利用荧光定量PCR分析lncRNA MIR205HG在肝癌组织与癌旁组织的表达差异;c,利用荧光定量PCR分析lncRNA MIR205HG在肝癌细胞和正常肝细胞中的表达差异;d、e,Hep 3B细胞经转染siRNA(20 nmol/L)处理24 h后,利用荧光定量PCR分别检测lncRNA MIR205HG和LNC01309的相对表达含量;f、g,Hep G2细胞中转染过表达质粒24 h后,利用荧光定量PCR分别检测LNC01309和miR-205的相对表达含量。
图 2 LNC01309与lncRNA MIR205HG的关系分析
表 1 利用NCBI BLAST比较LNC01309与lncRNA MIR205HG序列相似性
描述 种属 最高分 总分 查询覆盖值 E值 同源百分比 查询长度 登录号 Homo sapiens MIR205 host gene (MIR205HG), transcript variant 3, long non-coding RNA Homo sapiens 516 516 1 5×10-142 0.963 992 NR_145434.1 Homo sapiens MIR205 host gene (MIR205HG), transcript variant 1, long non-coding RNA Homo sapiens 516 516 1 5×10-142 0.963 895 NR_145437.1 Homo sapiens MIR205 host gene (MIR205HG), transcript variant 2, long non-coding RNA Homo sapiens 516 516 1 5×10-142 0.963 857 NR_145433.1 Homo sapiens MIR205 host gene (MIR205HG), transcript variant 4, long non-coding RNA Homo sapiens 516 516 1 5×10-142 0.963 940 NR_145435.1 Homo sapiens MIR205 host gene (MIR205HG), transcript variant 5, long non-coding RNA Homo sapiens 516 516 1 5×10-142 0.963 651 NR_145436.1 表 2 利用RBPmap和catRAPID在线预测能够与LNC01309结合的潜在靶标蛋白
蛋白质 序列位置 基序 K-mer Z评分 P值 CNOT4 129 gacaga gagaga 3.554 1.90×10-4 EIF4G2 50 cgccg cgccg 4.390 5.67×10-6 HNRNPA1 66 guaguagu guagcagc 3.361 3.88×10-4 MBNL1 70 gcgcagc cagcagc 3.903 4.75×10-5 RBM24 160 wgwgugd ggagugu 3.403 3.33×10-4 RBM38 163 kkguguk gugugcg 3.333 4.30×10-4 RBM6 37 ccacc ccacc 4.228 1.18×10-5 SRSF10 70 cagcag cagcag 4.513 3.20×10-6 -
[1] BHAN A, SOLEIMANI M, MANDAL SS. Long noncoding RNA and cancer: A new paradigm[J]. Cancer Res, 2017, 77(15): 3965-3981. DOI: 10.1158/0008-5472.CAN-16-2634. [2] PENG WX, KOIRALA P, MO YY. LncRNA-mediated regulation of cell signaling in cancer[J]. Oncogene, 2017, 36(41): 5661-5667. DOI: 10.1038/onc.2017.184. [3] MATTICK JS. The genetic signatures of noncoding RNAs[J]. PLoS Genet, 2009, 5(4): e1000459. DOI: 10.1371/journal.pgen.1000459. [4] KUMAR S, GONZALEZ EA, RAMESHWAR P, et al. Non-coding RNAs as mediators of epigenetic changes in malignancies[J]. Cancers (Basel), 2020, 12(12): 3657. DOI: 10.3390/cancers12123657. [5] MA L, BAJIC VB, ZHANG Z. On the classification of long non-coding RNAs[J]. RNA Biol, 2013, 10(6): 925-933. DOI: 10.4161/rna.24604. [6] MIYAMOTO M, MOTOOKA D, GOTOH K, et al. Performance comparison of second- and third-generation sequencers using a bacterial genome with two chromosomes[J]. BMC Genomics, 2014, 15: 699. DOI: 10.1186/1471-2164-15-699. [7] RHOADS A, AU KF. PacBio sequencing and its applications[J]. Genomics Proteomics Bioinformatics, 2015, 13(5): 278-289. DOI: 10.1016/j.gpb.2015.08.002. [8] XU J, ZHENG Y, PU S, et al. Third-generation sequencing found LncRNA associated with heat shock protein response to heat stress in Populus qiongdaoensis seedlings[J]. BMC Genomics, 2020, 21(1): 572. DOI: 10.1186/s12864-020-06979-z. [9] SHEN Y, LIANG W, LIN Y, et al. Single molecule real-time sequencing and RNA-seq unravel the role of long non-coding and circular RNA in the regulatory network during Nile tilapia (Oreochromis niloticus) infection with Streptococcus agalactiae[J]. Fish Shellfish Immunol, 2020, 104: 640-653. DOI: 10.1016/j.fsi.2020.06.015. [10] HUANG Z, ZHOU JK, PENG Y, et al. The role of long noncoding RNAs in hepatocellular carcinoma[J]. Mol Cancer, 2020, 19(1): 77. DOI: 10.1186/s12943-020-01188-4. [11] CHEN Y, LI Z, CHEN X, et al. Long non-coding RNAs: From disease code to drug role[J]. Acta Pharm Sin B, 2021, 11(2): 340-354. DOI: 10.1016/j.apsb.2020.10.001. [12] LI H, JI FX, XU XN, et al. Effects of long-chain non-coding RNA LINC00467 targeting miR-1247-5p on proliferation, migration and invasion of hepatocellular carcinoma cells[J]. Chin J Gerontol, 2021, 41(15): 3328-3333. DOI: 10.3969/j.issn.1005-9202.2021.15.048李豪, 姬发祥, 徐晓宁, 等. 长链非编码RNA LINC00467靶向miR-1247-5p调控肝癌细胞增殖、迁移及侵袭的影响[J]. 中国老年学杂志, 2021, 41(15): 3328-3333. DOI: 10.3969/j.issn.1005-9202.2021.15.048 [13] YUAN JH, YANG F, WANG F, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma[J]. Cancer Cell, 2014, 25(5): 666-681. DOI: 10.1016/j.ccr.2014.03.010. [14] PANZITT K, TSCHERNATSCH MM, GUELLY C, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA[J]. Gastroenterology, 2007, 132(1): 330-342. DOI: 10.1053/j.gastro.2006.08.026. [15] DU Y, KONG G, YOU X, et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18[J]. J Biol Chem, 2012, 287(31): 26302-26311. DOI: 10.1074/jbc.M112.342113. [16] GENG YJ, XIE SL, LI Q, et al. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression[J]. J Int Med Res, 2011, 39(6): 2119-2128. DOI: 10.1177/147323001103900608. [17] LI G, ZHANG H, WAN X, et al. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma[J]. Biomed Res Int, 2014, 2014: 780521. DOI: 10.1155/2014/780521. [18] MAASS PG, LUFT FC, BÄHRING S. Long non-coding RNA in health and disease[J]. J Mol Med (Berl), 2014, 92(4): 337-346. DOI: 10.1007/s00109-014-1131-8. [19] MENG YM, JIANG J, WANG YH, et al. Mechanism of long noncoding RNA encoded peptide in tumor[J]. Clin Misdiagn Misther, 2021, 34(9): 113-116. https://www.cnki.com.cn/Article/CJFDTOTAL-LCWZ202109024.htm孟一妹, 蒋晶, 王禹涵, 等. 长链非编码RNA编码肽在肿瘤中的作用机制[J]. 临床误诊误治, 2021, 34(9): 113-116. https://www.cnki.com.cn/Article/CJFDTOTAL-LCWZ202109024.htm [20] SCHAUKOWITCH K, KIM TK. Emerging epigenetic mechanisms of long non-coding RNAs[J]. Neuroscience, 2014, 264: 25-38. DOI: 10.1016/j.neuroscience.2013.12.009. [21] HUANG JZ, CHEN M, CHEN, et al. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth[J]. Mol Cell, 2017, 68(1): 171-184. DOI: 10.1016/j.molcel.2017.09.015. [22] LI XX, SHI L, ZHOU XJ, et al. The role of c-Myc-RBM38 loop in the growth suppression in breast cancer[J]. J Exp Clin Cancer Res, 2017, 36(1): 49. DOI: 10.1186/s13046-017-0521-5. [23] JIANG Y, XU E, ZHANG J, et al. The Rbm38-p63 feedback loop is critical for tumor suppression and longevity[J]. Oncogene, 2018, 37(21): 2863-2872. DOI: 10.1038/s41388-018-0176-5. [24] CHO SJ, ZHANG J, CHEN X. RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability[J]. Nucleic Acids Res, 2010, 38(7): 2256-2267. DOI: 10.1093/nar/gkp1229. [25] XU E, ZHANG J, CHEN X. MDM2 expression is repressed by the RNA-binding protein RNPC1 via mRNA stability[J]. Oncogene, 2013, 32(17): 2169-2178. DOI: 10.1038/onc.2012.238. [26] YAN W, ZHANG J, ZHANG Y, et al. p73 expression is regulated by RNPC1, a target of the p53 family, via mRNA stability[J]. Mol Cell Biol, 2012, 32(13): 2336-2348. DOI: 10.1128/MCB.00215-12. [27] SUN Z, YANG GS, SIMA H, et al. Expression of RBM5 and its effect on prognosis of hepatocellular carcinoma patients after hepatectomy[J/CD]. Chin J Hepat Surg (Electronic Edition), 2021, 10(1): 93-97. DOI: 10.3877/cma.j.issn.2095-3232.2021.01.020.孙喆, 杨广顺, 司马辉, 等. RBM5在肝细胞癌中的表达及其对肝切除术后患者预后的影响[J/CD]. 中华肝脏外科手术学电子杂志, 2021, 10(1): 93-97. DOI: 10.3877/cma.j.issn.2095-3232.2021.01.020. [28] YE J, LIANG R, BAI T, et al. RBM38 plays a tumor-suppressor role via stabilizing the p53-mdm2 loop function in hepatocellular carcinoma[J]. J Exp Clin Cancer Res, 2018, 37(1): 212. DOI: 10.1186/s13046-018-0852-x. 期刊类型引用(2)
1. 郭佳佳,张文果,王文秀,张晓丽,马徜徉. 血清Nox2、ATG7水平对新生儿窒息心肌损伤的评估价值. 安徽医药. 2024(09): 1791-1795 .  百度学术
百度学术2. 蔡卓玮,胡刚峰,章波. miR-203在乙型肝炎病毒相关肝细胞癌诊断及预后中的意义. 广西医科大学学报. 2022(11): 1773-1780 .  百度学术
百度学术其他类型引用(0)
-




 PDF下载 ( 5701 KB)
PDF下载 ( 5701 KB)

 下载:
下载:




 下载:
下载:









 百度学术
百度学术