肝细胞癌复发进程中DNA修复调节的蛋白质组学分析及验证
DOI: 10.12449/JCH240216
Proteomic analysis and validation of DNA repair regulation in the process of hepatocellular carcinoma recurrence
-
摘要:
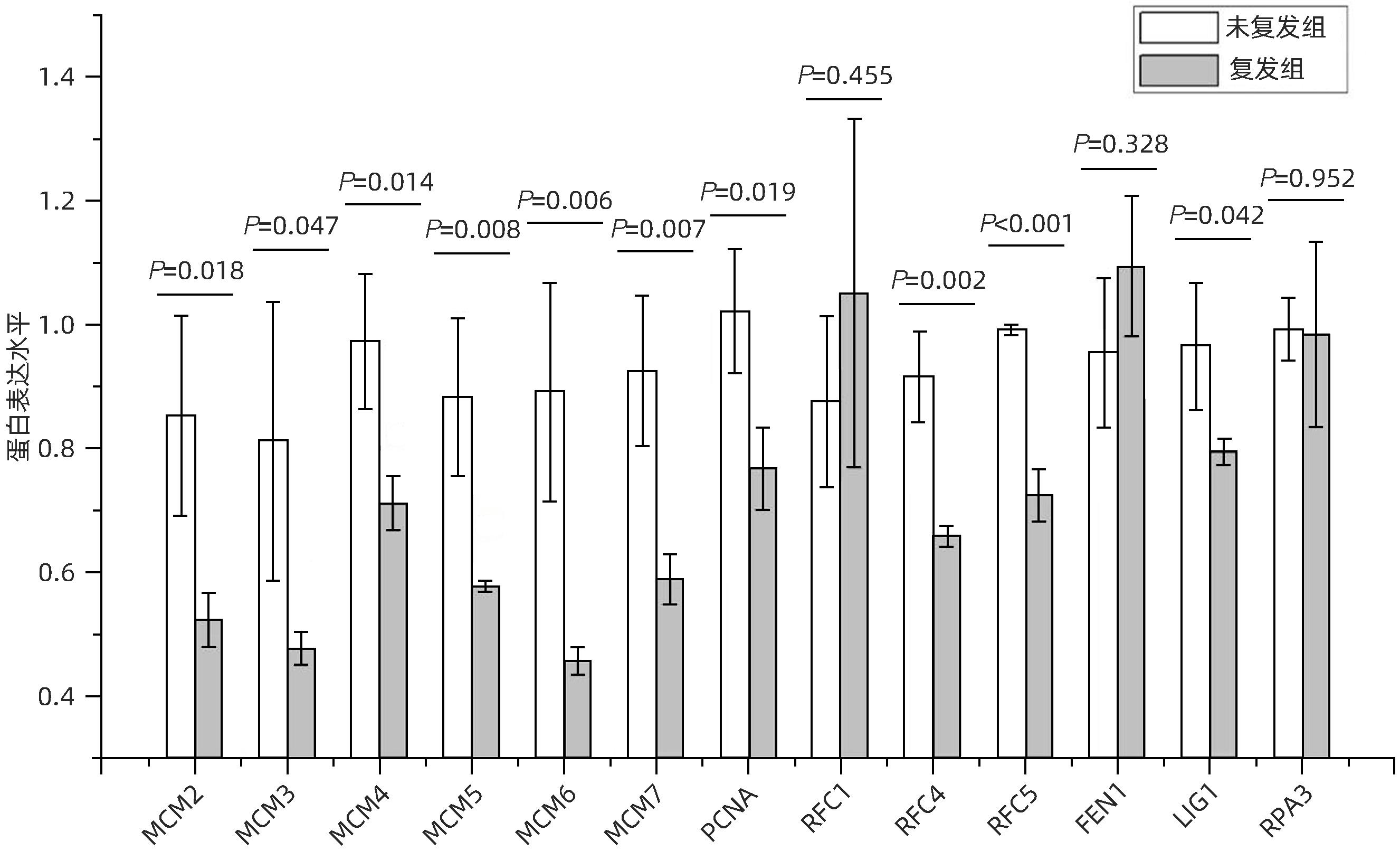
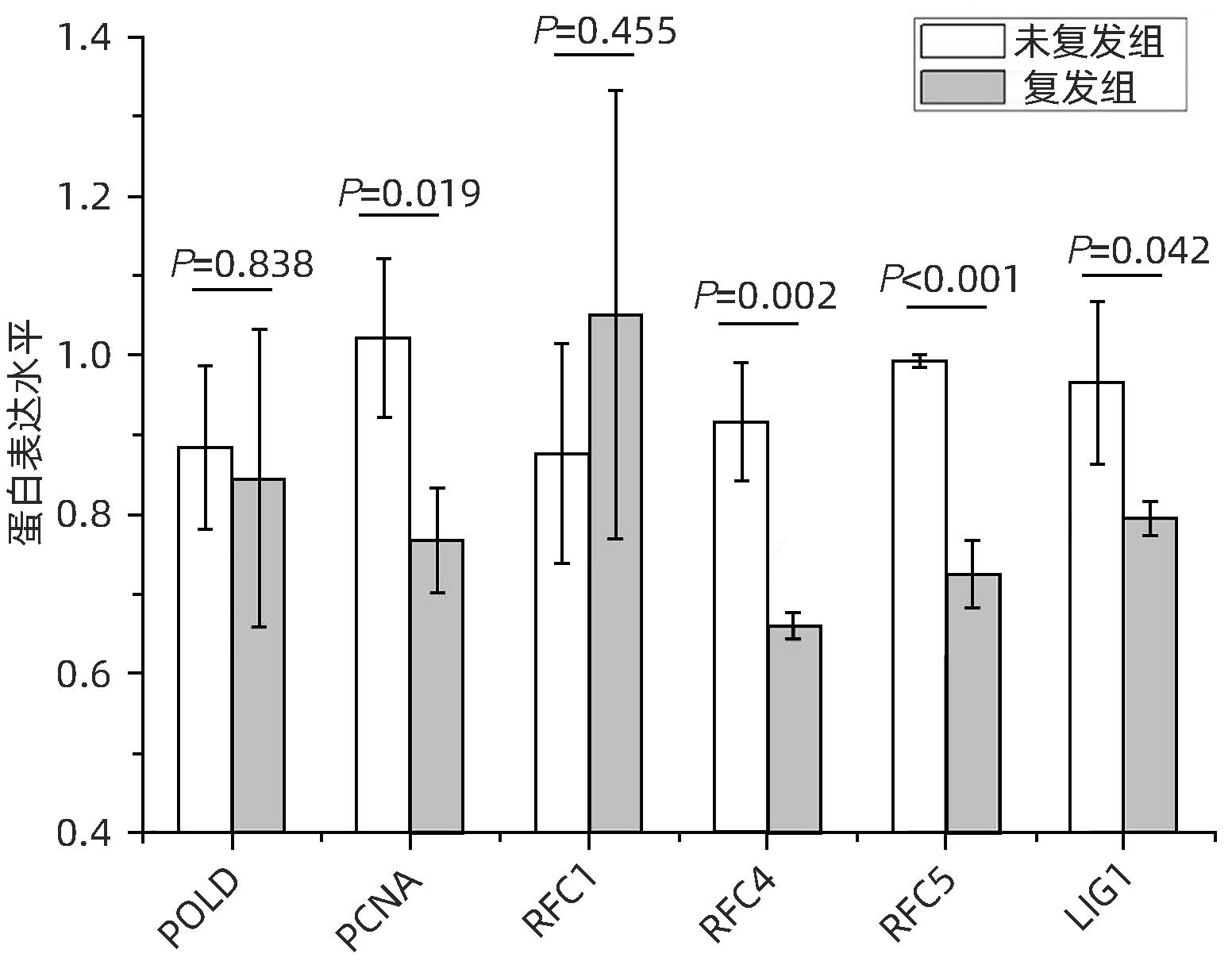
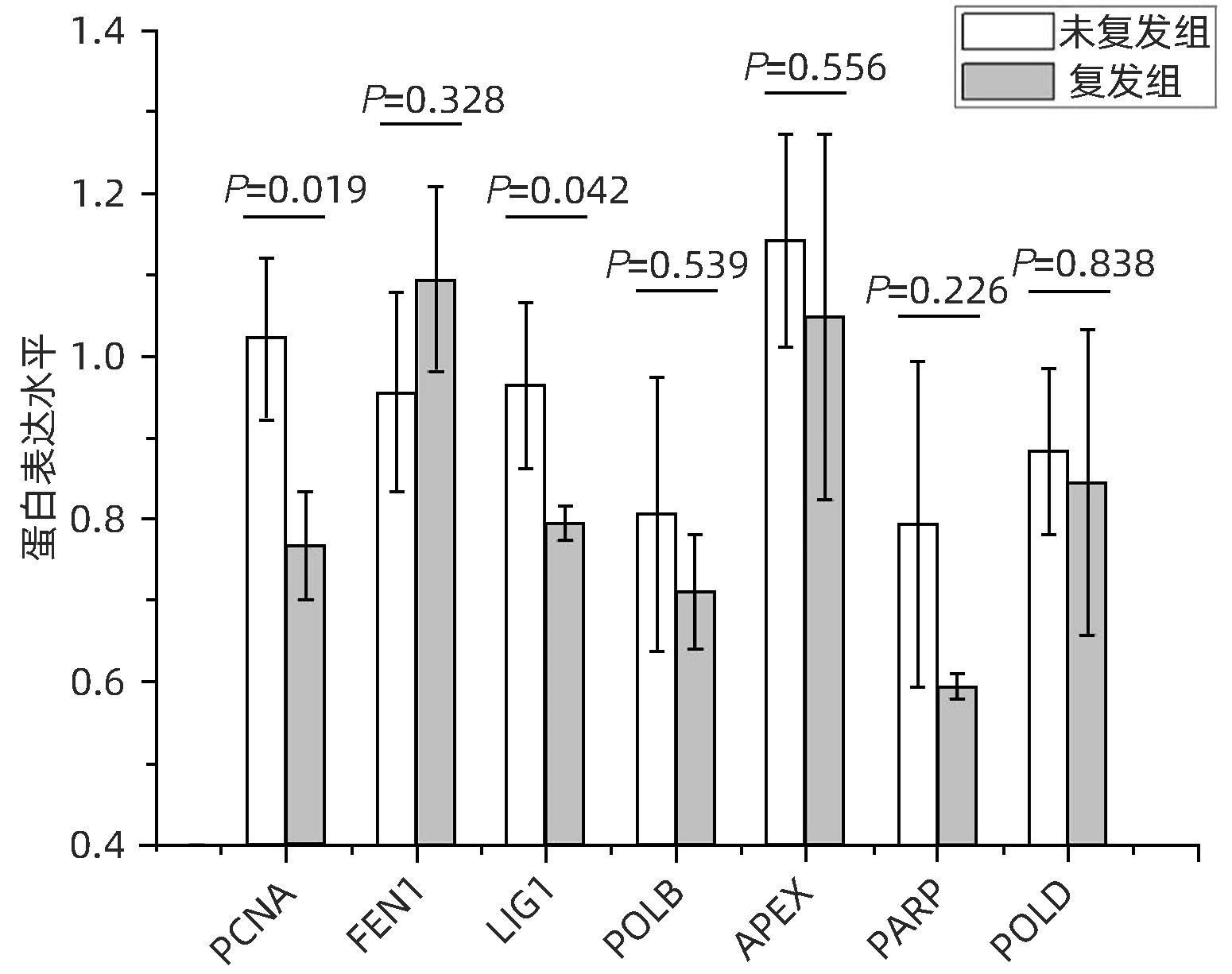
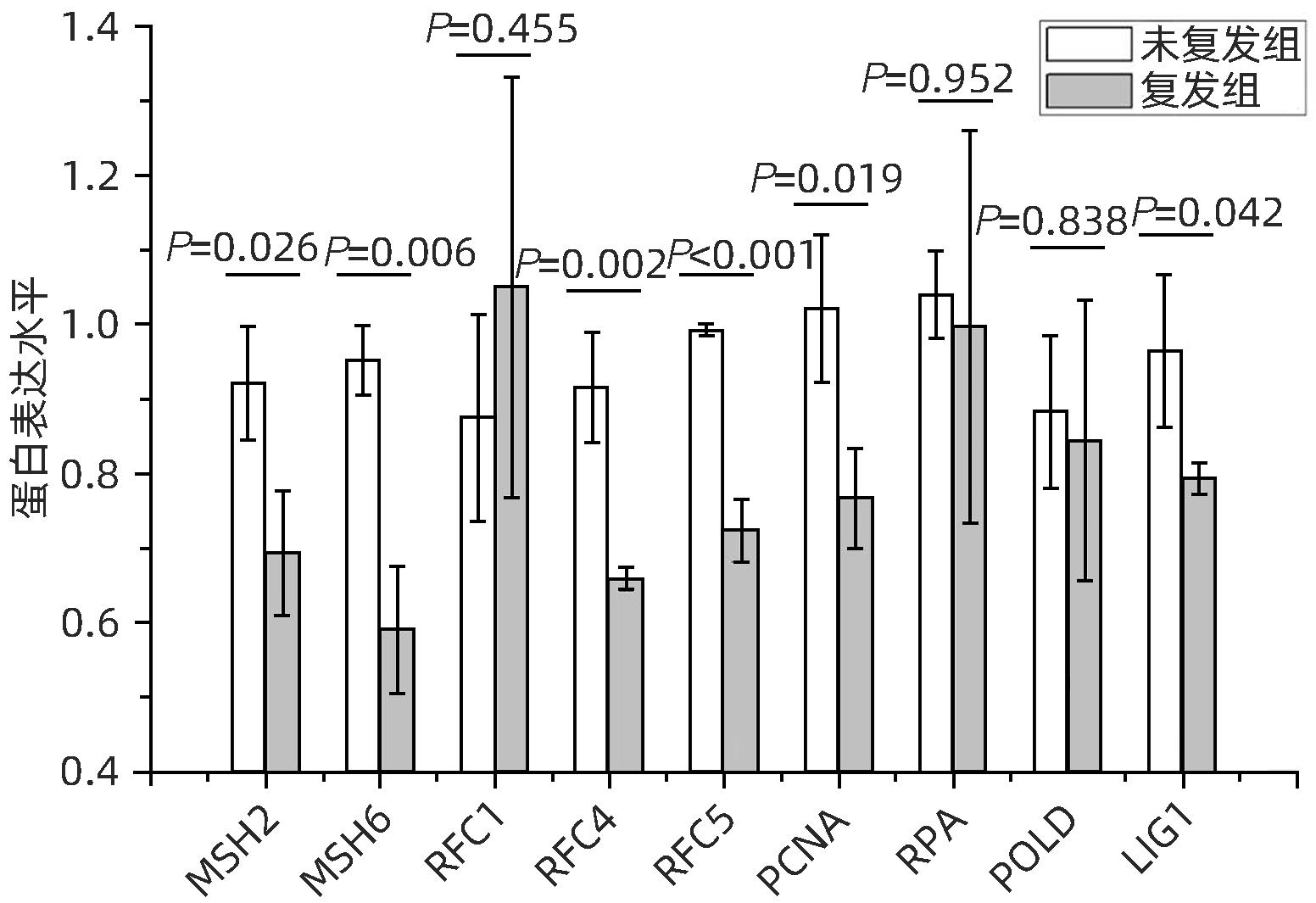
目的 分析肝细胞癌(HCC)复发进程中DNA修复调节的作用及机制。 方法 串联质量标签(TMT)标记的定量蛋白质组学方法分析HCC 2年内复发和5年预后良好的肝癌组织样本,分析富集在DNA复制、错配修复、碱基切除修复、核苷酸切除修复4条通路的蛋白表达差异,分析在HCC复发进程中起主要作用的调控通路及靶点,预测可能的调控机制。计量资料2组间比较采用成组t检验;多组间比较采用单因素方差分析,进一步两两比较采用LSD-t检验。 结果 真核生物复制复合体通路MCM2(P=0.018)、MCM3(P=0.047)、MCM4(P=0.014)、MCM5(P=0.008)、MCM6(P=0.006)、MCM7(P=0.007)、PCNA(P=0.019)、RFC4(P=0.002)、RFC5(P<0.001)、LIG1(P=0.042)蛋白表达均显著降低;核苷酸切除修复通路中PCNA(P=0.019)、RFC4(P=0.002)、RFC5(P<0.001)、LIG1(P=0.042)共4个蛋白表达水平均显著减少;碱基切除修复通路PCNA(P=0.019)和LIG1(P=0.042)在HCC复发组中均显著降低;错配修复富集通路中MSH2(P=0.026)、MSH6(P=0.006)、RFC4(P=0.002)、RFC5(P<0.001)、PCNA(P=0.019)、LIG1(P=0.042)共6个蛋白在肝癌复发组织中均显著减少。差异蛋白涉及MCM复合体、DNA聚合酶复合体ε、连接酶LIG1、长补丁碱基剪切修复复合体(long patch BER)、DNA错配修复蛋白复合体的重要组分。对DNA修复调节的重要差异蛋白进行临床样本验证分析,结果表明复发组中除MCM6表现出下降趋势外,MCM5(P=0.008)、MCM7(P=0.007)、RCF4(P=0.002)、RCF5(P<0.001)和MSH6(P=0.006)蛋白相对表达量均显著降低。 结论 HCC复发过程中,DNA修复进程中多个复合体蛋白组分存在显著减少或缺失。 Abstract:Objective To investigate the role and mechanism of DNA repair regulation in the process of hepatocellular carcinoma (HCC) recurrence. Methods HCC tissue samples were collected from the patients with recurrence within two years or the patients with a good prognosis after 5 years, and the Tandem Mass Tag-labeled quantification proteomic study was used to analyze the differentially expressed proteins enriched in the four pathways of DNA replication, mismatch repair, base excision repair, and nucleotide excision repair, and the regulatory pathways and targets that play a key role in the process of HCC recurrence were analyzed to predict the possible regulatory mechanisms. The independent samples t-test was used for comparison of continuous data between two groups; a one-way analysis of variance was used for comparison between multiple groups, and the least significant difference t-test was used for further comparison between two groups. Results For the eukaryotic replication complex pathway, there were significant reductions in the protein expression levels of MCM2 (P=0.018), MCM3 (P=0.047), MCM4 (P=0.014), MCM5 (P=0.008), MCM6 (P=0.006), MCM7 (P=0.007), PCNA (P=0.019), RFC4 (P=0.002), RFC5 (P<0.001), and LIG1 (P=0.042); for the nucleotide excision repair pathway, there were significant reductions in the protein expression levels of PCNA (P=0.019), RFC4 (P=0.002), RFC5 (P<0.001), and LIG1 (P=0.042); for the base excision repair pathway, there were significant reductions in the protein expression levels of PCNA (P=0.019) and LIG1 (P=0.042) in the HCC recurrence group; for the mismatch repair pathway, there were significant reductions in the protein expression levels of MSH2 (P=0.026), MSH6 (P=0.006), RFC4 (P=0.002), RFC5 (P<0.001), PCNA (P=0.019), and LIG1 (P=0.042) in recurrent HCC tissue. The differentially expressed proteins were involved in the important components of MCM complex, DNA polymerase complex, ligase LIG1, long patch base shear repair complex (long patch BER), and DNA mismatch repair protein complex. The clinical sample validation analysis of important differentially expressed proteins regulated by DNA repair showed that except for MCM6 with a trend of reduction, the recurrence group also had significant reductions in the relative protein expression levels of MCM5 (P=0.008), MCM7 (P=0.007), RCF4 (P=0.002), RCF5 (P<0.001), and MSH6 (P=0.006). Conclusion There are significant reductions or deletions of multiple complex protein components in the process of DNA repair during HCC recurrence. -
Key words:
- Carcinoma, Hepatocellular /
- Neoplasm Recurrence, Local /
- DNA Repair /
- Proteomics
-
注: HCC/AllTumor,通过HCC与所有肿瘤的比较,红色/蓝色表示log2量表中的正/负倍比差异(TCGA数据);HCC/AllAdjacent,通过HCC与所有癌旁组织进行比较,红色/蓝色表示log2量表中的正/负倍比差异(TCGA数据);HCC/Adjacent,通过HCC与癌旁组织进行比较,红色/蓝色表示log2量表中的正/负倍比差异(HCCDB数据);Liver/OtherNormal,通过正常肝脏与其他正常组织的比较,红色/蓝色表示log2量表中的正/负倍比差异(GTEx和TCGA数据)。
图 7 差异蛋白在不同组织间的差异表达四维图谱
Figure 7. 4-D map of the differential expression of the differential proteins among different tissues
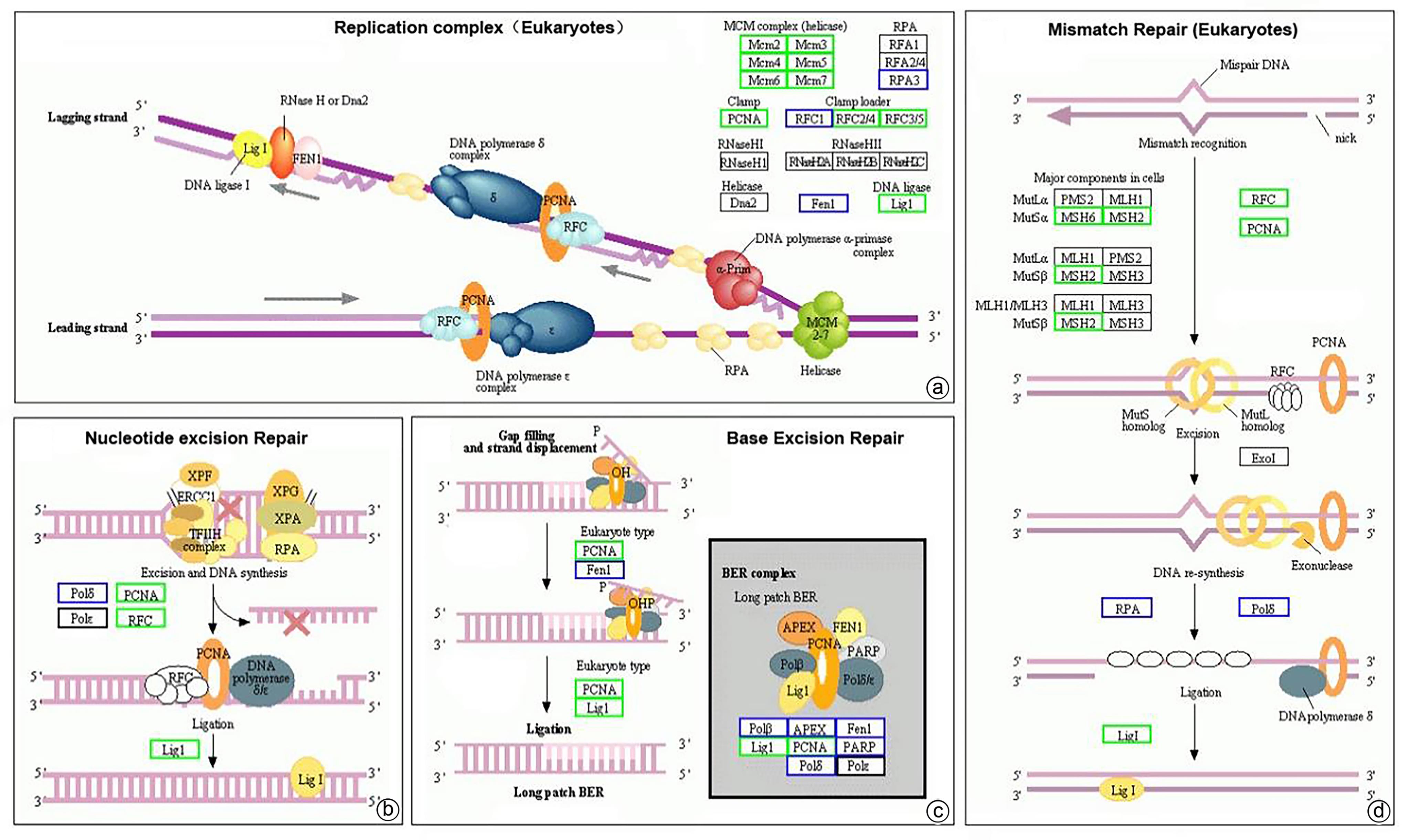
注: a,复制复合体(真核生物)富集通路图;b,核苷酸切除修复富集通路图;c,碱基剪切修复富集通路图;d,错配修复(真核生物)富集通路图。绿色方框代表HCC复发组中该蛋白表达量显著减少(P<0.05);蓝色方框代表HCC复发组中该蛋白表达量差异无统计学意义(P>0.05);黑色方框代表在该研究中未检出相关蛋白表达。
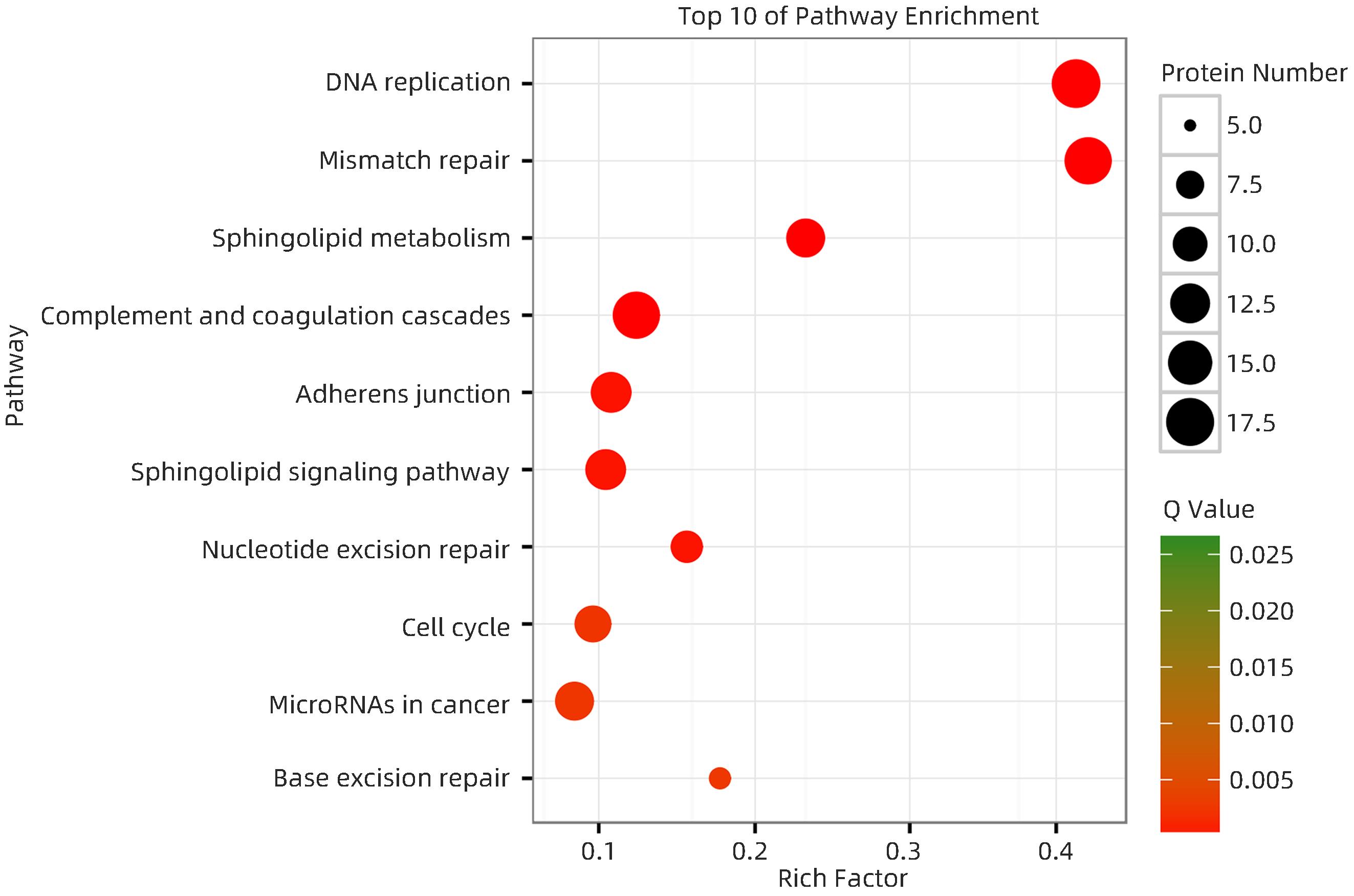
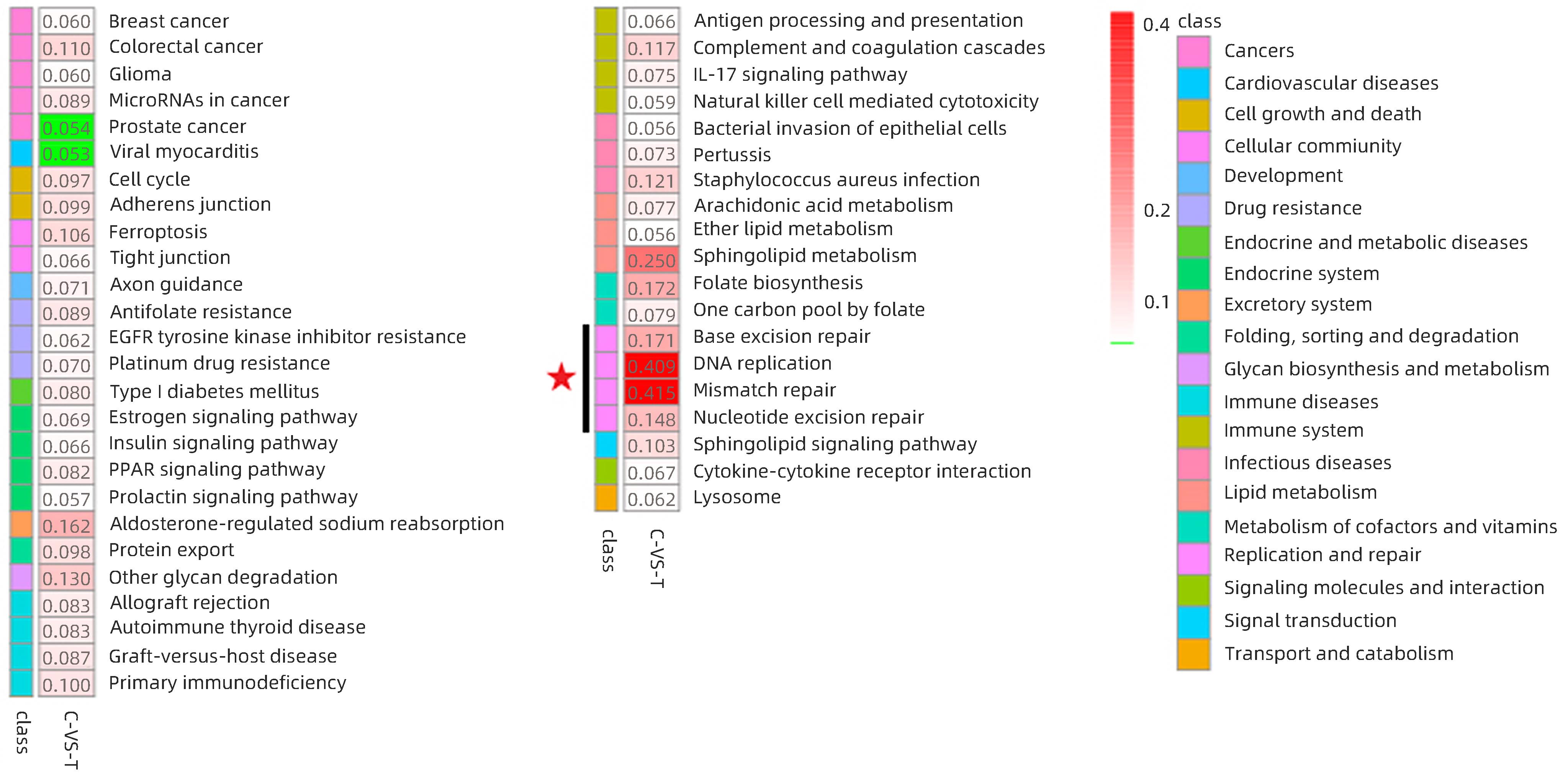
图 8 HCC复发组与未复发组差异蛋白富集通路图(差异分析基于KEGG pathway数据库)
Figure 8. Differential protein enrichment pathway map in the HCC recurrent and unrecurrent groups(Differential analysis was based on the KEGG pathway database)
-
[1] HU ZQ, YIN YF, JIANG J, et al. Exosomal miR-142-3p secreted by hepatitis B virus(HBV)-hepatocellular carcinoma(HCC) cells promotes ferroptosis of M1-type macrophages through SLC3A2 and the mechanism of HCC progression[J]. J Gastrointest Oncol, 2022, 13( 2): 754- 767. DOI: 10.21037/jgo-21-916. [2] KATO K, FUKAI M, HATANAKA KC, et al. Versican secreted by cancer-associated fibroblasts is a poor prognostic factor in hepatocellular carcinoma[J]. Ann Surg Oncol, 2022, 29( 11): 7135- 7146. DOI: 10.1245/s10434-022-11862-0. [3] PANG SJ, SHI Y, XU DP, et al. Screening of hepatocellular carcinoma patients with high risk of early recurrence after radical hepatectomy using a nomogram model based on the γ-glutamyl transpeptidase-to-albumin ratio[J]. J Gastrointest Surg, 2022, 26( 8): 1- 9. DOI: 10.1007/s11605-022-05326-9. [4] GOMEZ-QUIROZ LE, ROMAN S. Influence of genetic and environmental risk factors in the development of hepatocellular carcinoma in Mexico[J]. Ann Hepatol, 2022, 27( Suppl 1): 100649. DOI: 10.1016/j.aohep.2021.100649. [5] LUO MJ, ZHAO YX, WANG YD, et al. Comparative proteomics of contrasting maize genotypes provides insights into salt-stress tolerance mechanisms[J]. J Proteome Res, 2018, 17( 1): 141- 153. DOI: 10.1021/acs.jproteome.7b00455. [6] SHRESTHA P, KIM MS, ELBASANI E, et al. Prediction of trehalose-metabolic pathway and comparative analysis of KEGG, MetaCyc, and RAST databases based on complete genome of Variovorax sp. PAMC28711[J]. BMC Genom Data, 2022, 23( 1): 4. DOI: 10.1186/s12863-021-01020-y. [7] MURANUSHI R, ARAKI K, YOKOBORI T, et al. High membrane expression of CMTM6 in hepatocellular carcinoma is associated with tumor recurrence[J]. Cancer Sci, 2021, 112( 8): 3314- 3323. DOI: 10.1111/cas.15004. [8] ZHONG F, XIE C, PENG X, et al. A commentary on‘Laparoscopic versus open repeat hepatectomy for recurrent hepatocellular carcinoma: A systematic review and meta-analysis of propensity score-matched cohort studies’[J]. Int J Surg, 2023, 109( 9): 2821- 2822. DOI: 10.1097/js9.0000000000000517. [9] BAI YP, SHA JJ, OKUI T, et al. The epithelial-mesenchymal transition influences the resistance of oral squamous cell carcinoma to monoclonal antibodies via its effect on energy homeostasis and the tumor microenvironment[J]. Cancers, 2021, 13( 23): 5905. DOI: 10.3390/cancers13235905. [10] HUANG DP, LIAO MM, TONG JJ, et al. Construction of a genome instability-derived lncRNA-based risk scoring system for the prognosis of hepatocellular carcinoma[J]. Aging, 2021, 13( 22): 24621- 24639. DOI: 10.18632/aging.203698. [11] CHEN Y, HUANG MJ, ZHU JK, et al. Identification of a DNA damage response and repair-related gene-pair signature for prognosis stratification analysis in hepatocellular carcinoma[J]. Front Pharmacol, 2022, 13: 857060. DOI: 10.3389/fphar.2022.857060. [12] BARCENA-VARELA M, LUJAMBIO A. A novel long noncoding RNA finetunes the DNA damage response in hepatocellular carcinoma[J]. Cancer Res, 2021, 81( 19): 4899- 4900. DOI: 10.1158/0008-5472.CAN-21-2776. [13] WANG ZY, WANG XX, RONG ZH, et al. LncRNA LINC01134 contributes to radioresistance in hepatocellular carcinoma by regulating DNA damage response via MAPK signaling pathway[J]. Front Pharmacol, 2022, 12: 791889. DOI: 10.3389/fphar.2021.791889. [14] LIU HH, YAN YC, CHEN RB, et al. Integrated nomogram based on five stage-related genes and TNM stage to predict 1-year recurrence in hepatocellular carcinoma[J]. Cancer Cell Int, 2020, 20: 140. DOI: 10.1186/s12935-020-01216-9. [15] MARSHALL AE, RUSHBROOK SM, VOWLER SL, et al. Tumor recurrence following liver transplantation for hepatocellular carcinoma: Role of tumor proliferation status[J]. Liver Transpl, 2010, 16( 3): 279- 288. DOI: 10.1002/lt.21993. [16] LIAO XW, LIU XG, YANG CK, et al. Distinct diagnostic and prognostic values of minichromosome maintenance gene expression in patients with hepatocellular carcinoma[J]. J Cancer, 2018, 9( 13): 2357- 2373. DOI: 10.7150/jca.25221. [17] THUL PJ, LINDSKOG C. The human protein atlas: A spatial map of the human proteome[J]. Protein Sci, 2018, 27( 1): 233- 244. DOI: 10.1002/pro.3307. -



 PDF下载 ( 2144 KB)
PDF下载 ( 2144 KB)

 下载:
下载: