Changes in intestinal flora in patients with extrahepatic cholangiocarcinoma
-
摘要:
目的 研究肝外胆管癌(ECC)患者肠道菌群变化及影响因素。 方法 收集2020年1月—2020年12月于郑州大学第一附属医院肝胆胰外科住院治疗的16例ECC患者(ECC组)的粪便样本,采用菌群16S rRNA绝对定量测序,与胆总管结石患者(CBDS组)(20例)及健康对照(Normal组)(10例)进行比较,分析3组肠道菌群差异及与临床指标的相关性。符合正态分布且满足方差齐性的2组间比较采用t检验,3组之间比较采用单因素方差分析;非正态分布3组之间比较采用Kruskal-Wallis H秩和检验,组间比较采用Nemenyi检验。计数资料组间比较采用χ2检验或Fisher确切概率法,相关性检验采用Spearman相关性分析。 结果 ECC组TBil和DBil明显高于CBDS组和Normal组(P值均<0.05)。多样性分析显示Observed species、Chao1指数、Shannon指数在3组之间差异无统计学意义(P值均>0.05);而Shannon指数和Simpson指数在3组之间差异有统计学意义,ECC组物种多样性与Normal组相似,但高于CBDS组(P<0.05),CBDS组高于Normal组(P<0.05)。多样性分析显示ECC组菌群结构明显不同于Normal组和CBDS组(P<0.05)。菌群组成差异性分析表明,Prevotella、Lactobacillus、Megasphaera、Sutterella在ECC组中明显富集。相关分析显示,Prevotella与抗生素、抑酸药和保肝药的使用呈负相关,Lactobacillus、Megasphaera、Sutterella与TBil和DBil呈正相关。 结论 ECC组患者肠道菌群发生了明显改变,且菌群改变与ECC患者肝功能和药物的使用密切相关。 Abstract:Objective To investigate the changes in intestinal flora in patients with extrahepatic cholangiocarcinoma (ECC) and related influencing factors. Methods Fecal samples were collected from 16 patients with ECC who were hospitalized and treated in Department of Hepatobiliary and Pancreatic Surgery, The First Affiliated Hospital of Zhengzhou University, from January to December 2020, and absolute quantitative bacterial 16S rRNA was used for sequencing. A comparison was made with 20 patients with common bile duct stones (CBDS group) and 10 healthy controls (normal group), and the three groups were compared in terms of the differences in intestinal flora and the association with clinical indices. A one-way analysis of variance was used for comparison of normally distributed data with homogeneity of variance between the three groups, the t-test was used for comparison between two groups; the Kruskal-Wallis H test was used for comparison of non-normally distributed data between the three groups, and the Nemenyi test was used for comparison between groups. The chi-square test or the Fisher's exact test were used for comparison of categorical data between the three groups, and a Spearman correlation analysis was used to investigate correlation. Results The ECC group had significantly higher levels of total bilirubin (TBil) and direct bilirubin (DBil) than the CBDS group and the normal group. The α diversity analysis showed that there were no significant differences in observed species, Chao1 Index, and Shannon Index between the three groups (all P > 0.05), while there were significant differences in Shannon Index and Simpson Index between the three groups. The ECC group had a similar species diversity to the normal group and a significantly greater species diversity than the CBDS group (P < 0.05), and the CBDS group had a significantly greater species diversity than the normal group (P < 0.05). The β diversity analysis showed that the structure of intestinal flora in the ECC group was significantly different from that in the normal group and the CBDS group (P < 0.05). The analysis of the difference in bacterial composition showed that Prevotella, Lactobacillus, Megasphaera, and Sutterella were significantly enriched in the ECC group. The correlation analysis showed that Prevotella was negatively correlated with the use of antibiotics, acid inhibitors, and liver-protecting drugs, and Lactobacillus, Megasphaera, and Sutterella were positively correlated with TBil and DBil. Conclusion There is a significant change in intestinal flora in patients with ECC, which is closely associated with liver function and the use of drugs. -
胆管癌是一种源于胆道上皮的恶性肿瘤,根据解剖位置不同,胆管癌分为肝内胆管癌和肝外胆管癌(extrahepatic cholangiocarcinoma, ECC)。近几十年,全球ECC的发病率不断上升[1]。寄生虫感染、原发性硬化性胆管炎(PSC)、胆管囊肿和结石是ECC发生的危险因素[2]。肠道菌群占人体所有共生微生物的绝大多数,其种类超过1000种,细胞总数约1014,是人体自身细胞数量的10倍[3],对机体代谢、免疫等功能具有重要的调节作用[4]。体内外各种有害刺激因素都会导致肠道菌群发生紊乱,对机体产生严重损害。肠道菌群紊乱与慢性肝病、肝硬化和肝癌的形成发展密切相关,被形象地称为“肠-肝”轴[5-6]。本研究拟通过菌群16S rRNA绝对定量测序分析ECC患者肠道菌群变化,从菌群角度为ECC诊断和治疗提供理论依据。
1. 资料与方法
1.1 研究对象
2020年1月—2020年12月于郑州大学第一附属医院肝胆胰外科住院治疗的ECC患者16例(ECC组),诊断符合2020年美国国立综合癌症网络肝胆肿瘤临床实践指南(V1版)[7]标准;胆总管结石(common bile duct stones, CBDS)患者20例(CBDS组),诊断符合2017年胆总管结石管理指南[8];同期在本院体检的健康成人10例(Normal组)。排除标准:(1)ECC组排除肝内胆管癌、肝癌患者;确诊为ECC但是以往接受过手术或介入治疗的患者。(2)CBDS组排除胆囊结石和肝内胆管结石患者。(3)有其他恶性肿瘤;信息不完全;滥用酒精;既往消化道疾病史或消化道手术史;免疫系统疾病以及其他与肠道菌群相关的疾病(如炎症性肠病、糖尿病)。
1.2 样品采集及DNA提取
晨起后排便于无菌容器,使用一次性采集勺取适量样品于无菌采集管中,置于冰盒中迅速送至实验室,分装后置于-80 ℃冰箱保存,保存时间小于1个月。按QIAampⓇ Fast DNA Stool Mini Kit(Germany, Qiagen)说明书提取粪便菌群总DNA,NanoDrop2000(USA, Thermo Fisher Scientific)检测DNA浓度,琼脂糖凝胶电泳检测DNA完整性。
1.3 16S rRNA可变区扩增及测序
选取16S rRNA V3-V4可变区进行PCR扩增,引物为F(5′-CCTACGGGNGGCWGCAG-3′) 和R(5′-GACTACHVGGGTATCTAATCC-3′)。利用带有Index序列的引物,通过高保真PCR向文库末端引入特异性标签序列,经纯化后(Agencourt AMPure XP,USA),得到原始文库。再利用Invitrogen对文库进行精确定量(Qubit 3.0,USA),根据不同样本的测序通量要求,按相应摩尔比混合样本。使用生物分析仪(Agilent 2100,USA)评估DNA文库的质量和检测插入物的大小,所有样品插入物的大小在120~200 bp。合格后通过使用Illumina NovaSeq(Illumina,USA)台式序列仪,用2×250 bp配对末端方法进行16S rRNA基因扩增子的测序。
1.4 数据信息分析
首先对原始下机数据进行质控和过滤(QIIME2,2019.10),使用优化的有效序列进行ASVs(Amplicon sequence variants)分类分析。标记spike-in序列,并统计各个样本spike-in序列比例。根据每个样本spike-in序列数和拷贝数信息制作标准曲线方程,计算得到每个样本中各个ASVs的绝对拷贝数(不含spike-in序列)。ASVs的分类学物种注释,默认使用RDP数据库(https://rrndb.umms.med.umich.edu/)。Alpha多样性分析使用MOTHUR软件进行,群落组成分析使用MOTHUR和R软件完成。Beta多样性分析和组间显著差异的物种,主要使用R和Lefse软件完成。差异菌群与环境因子的相关性分析采用Spearman分析。
1.5 伦理学审查
本研究经郑州大学第一附属医院伦理委员会批准,批号:2020-KY-369,患者及家属均签署知情同意书。
1.6 统计学方法
应用SPSS 26.0软件进行数据分析处理,符合正态分布计量资料以x±s表示,不符合正态分布计量资料用M(P25~P75)表示;符合正态分布且满足方差齐性的2组间比较采用t检验,3组之间比较采用单因素方差分析;非正态分布3组之间比较采用Kruskal-Wallis H秩和检验,组间比较采用Nemenyi检验。计数资料组间比较采用χ2检验或Fisher确切概率法。差异物种与临床指标及药物使用的相关性采用Spearmans相关性分析。P<0.05为差异有统计学意义。
2. 结果
2.1 一般资料
ECC组TBil、DBil、ALP和GGT明显高于CBDS组(P值均<0.05)。在药物使用方面,ECC组使用过或正在使用抑酸药、保肝药和抗生素的例数分别为4例、4例、3例,与CBDS组(7例、10例、9例)比较,差异均无统计学意义(P值均>0.05),ECC组使用抑酸药、保肝药和抗生素的天数与CBDS组相比差异均无统计学意义(P值均>0.05)。另外,ECC组有2例接受过经皮肝穿刺胆道引流术(percutaneous transhepatic cholangial drainage, PTCD)减黄。ECC组与Normal组比较,肝功能指标差异均有统计学意义(P值均<0.05)(表 1)。
表 1 3组一般资料比较项目 ECC组(n=16) CBDS组(n=20) Normal组(n=10) 统计值 P值 男/女(例) 7/9 7/13 5/5 χ2=0.679 0.891 年龄(岁) 61.38±10.69 60.25±16.07 53.20±11.55 F=1.257 0.295 TBil(mol/L) 176.59(63.56~324.11) 29.24(14.74~113.73)1) 14.10(12.23~15.78)1) χ2=22.471 <0.001 DBil(mol/L) 150.16(55.11~278.45) 20.79(10.40~101.64)1) 6.96(6.18~8.35)1) χ2=24.575 <0.001 ALP(U/L) 268.65(200.25~437.00) 155.00(67.75~319.00)1) 77.50(68.25~89.00)1) χ2=18.447 <0.001 GGT(U/L) 379.00(226.50~774.75) 188.5(83.00~411.5)1) 33.00(21.00~39.50)1) χ2=23.311 <0.001 ALT(U/L) 80.50(48.25~102.98) 59.00(39.53~123.75) 21.50(18.75~29.00)1) χ2=17.767 <0.001 AST(U/L) 67.00(41.50~100.75) 46.00(27.25~85.00) 23.00(18.25~29.25)1) χ2=15.188 0.001 使用抑酸药天数(d) 13.00±10.67 16.54±8.25 t=0.290 0.885 使用保肝药天数(d) 12.00±5.38 6.60±9.11 t=0.843 0.724 使用抗生素天数(d) 12.33±18.06 14.77±2.34 t=0.621 0.967 PTCD治疗(例) 2 0 0.190 注: 与ECC组比较, 1)P<0.05。 2.2 测序深度评估
分别从样本中随机抽取出一部分序列,以抽到的序列数与它们所能代表ASVs的数目构建稀释性曲线。可见曲线随着抽取序列数的增加而趋于平缓,说明样本的测序数据量合理, 足以覆盖大多数微生物(图 1)。
2.3 肠道菌群多样性分析
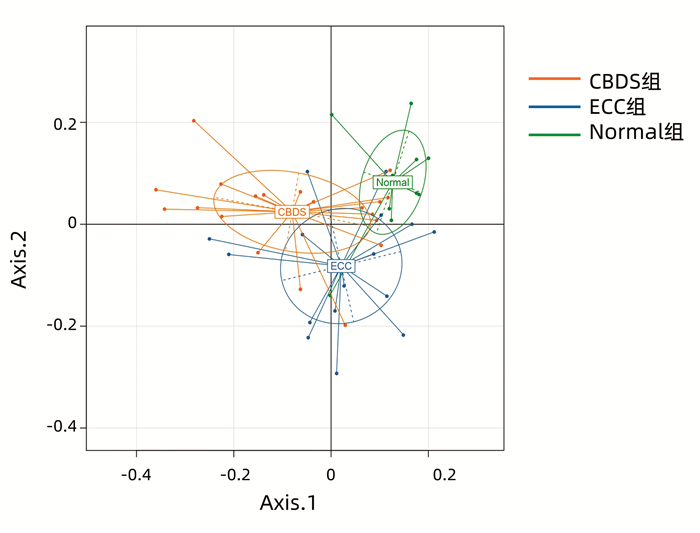
多样性是描述样本物种总数和多样性的指标,主要包括Observed species、Chao1指数、Shannon指数、ACE指数和Simpson指数。Observed species显示3组间物种总数未发生明显变化,Chao1指数和ACE指数显示3组间物种多样性无明显差异,而Shannon指数和Simpson指数显示3组物种多样性有明显差异,ECC组物种多样性与Normal组相似,但高于CBDS组;CBDS组高于Normal组(表 2)。多样性反映的是组间菌群组成差异,本文采用主坐标分析(PCoA)来直观显示,如图 2所示,3组样本区域尽管在X轴与Y轴上存在一定重合,但3组中心存在距离,因此3组间菌群构成存在差异。
表 2 3组间菌群多样性的比较指数 ECC组(n=16) CBDS组(n=20) Normal组(n=10) F值 P值 Observed species 243±51 211±87 240±68 0.420 0.403 Chao1 244.43±51.59 213.24±88.55 240.98±67.62 0.420 0.421 Shannon 3.50±0.441) 2.48±0.99 3.37±0.381) 0.007 0.002 ACE 244.69±51.78 213.53±88.37 241.39±67.65 0.420 0.407 Simpson 0.072±0.0411) 0.259±0.210 0.091±0.1761) 0.002 <0.001 注: 与CBDS组比较,1)P<0.05。 2.4 差异菌属鉴定
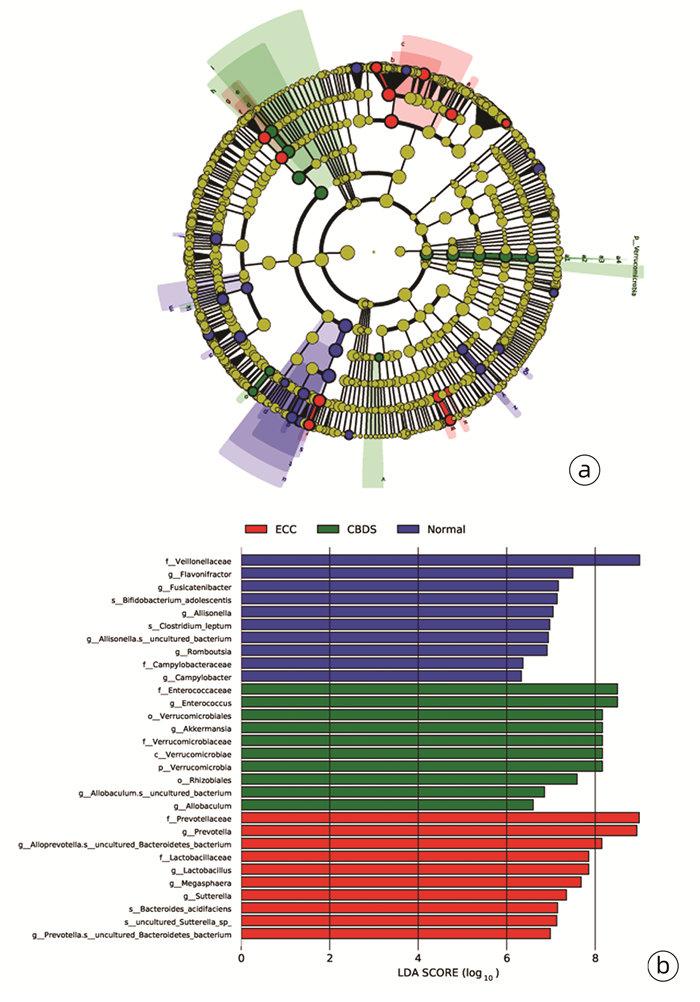
3组差异菌属鉴定如图 3所示:Citrobacter在Normal组中的含量显著高于ECC组和CBDS组(P<0.001)。Paraprevotella和Flavonifractor在Normal组中的含量显著高于ECC组和CBDS组(P值均<0.01)。此外,在Normal组中Bilophila、Bacteroides、Fusicatenibacter、Megamonas、Alistipes和Anaerofilum的含量增加,与ECC组和CBDS组相比,差异均有统计学意义(P值均<0.05)。Allobaculum在CBDS组中的含量与ECC组相比, 差异有统计学意义(P<0.01)。CBDS组和ECC组Sutterella的含量与Normal组相比,差异均有统计学意义(P值均<0.05)。
2.5 3组菌群组成差异性分析
线性判别分析(LEfSe)显示,在门水平上,ECC组和Normal组中无代表性的菌门,而Verrucomicrobia在CBDS组明显富集。在科水平上,Lactobacillaceae和Prevotellaceae在ECC组中明显增多,Enterococcaceae和Verrucomicrobiaceae在CBDS组中明显增多,而在Normal组中Veillonellaceae和Campylobacteraceae明显富集。在属类水平上,ECC组中显著差异的菌属是Prevotella、Lactobacillus、Megasphaera、Sutterella、Alloprevotella.s__uncultured_Bacteroidetes_bacterium和Prevotella.s__uncultured_Bacteroidetes_bacterium,Normal组中显著差异的菌属是Flavonifractor、Fusicatenibacter、Allisonella、Allisonella.s__uncultured_bacterium、Campylobacter和Romboutsia。除此之外,Enterococcus、Akkermansia、Allobaculum.s__uncultured_bacterium和Allobaculum是CBDS组中显著差异的菌属(图 4)。
2.6 菌群与临床指标的相关性分析
Prevotella与抗生素、抑酸药和保肝药的使用呈负相关。Lactobacillus、Megasphaera、Sutterella与TBil和DBil呈正相关,此外,Lactobacillus还与AST、ALT、ALP、GGT和年龄呈正相关,Megasphaera和ALP呈正相关,Sutterella与AST呈正相关(附件1)。
3. 讨论
胆管癌是一种侵袭性高且预后差的肿瘤,根治性手术切除是首选治疗方法,但多数患者就诊时已是晚期,能根治性切除的仅占35%[1]。ECC是胆管癌最常见的类型,占比约75%[9],5年生存率在20%~30%[10]。最近研究[11]表明胆管癌患者肠道菌群明显发生紊乱,但对ECC患者肠道菌群变化还未见相关研究。
本研究结果表明无论是多样性分析或菌群差异分析,均提示ECC菌群构成发生了明显改变。首先,ECC组的多样性与Normal组相似,却高于CBDS组,Chen等[12]同样发现远端胆管癌患者肠道菌群多样性明显高于胆总管结石患者。其次在多样性的分析中,ECC组的菌群结构与CBDS组和Noaml组明显不同,这与Lee等[11]和Avilés-Jiménez等[13]研究的结果一致。此外,与ECC组和Normal组相比,CBDS组物种多样性明显减少,有研究[14]报道称与健康人群相比,胆总管结石患者肠道菌群多样性明显减低,且肠道菌群紊乱与胆总管结石形成密切相关,其机制可能是通过影响胆汁酸和胆固醇代谢导致。
Proteobacteria大多为条件性致病菌,是脂多糖主要生成来源,与炎症反应密切相关,且大鼠中脂多糖暴露会导致快速的肝损伤[15]。本研究结果显示,ECC组中Sutterella(Proteobacteria的成员)明显富集且Sutterella的占比与TBil、DBil呈正相关。说明炎症反应在ECC的疾病演化中仍扮演重要角色,但炎症反应程度可能较CBDS轻缓,持续时间长。Sutterella对人胆汁酸具有较强抵抗力,这可能是其在胆道和肠道中存活的原因[15]。此外,Sutterella在十二指肠中含量最高,从十二指肠至结肠梯度递减,具有轻度的促炎能力[16-17]。
Bacteroidetes大约有7000个不同物种,是最大的革兰阴性菌门类[18]。Prevotella是其主要菌属[19],笔者发现Prevotella在ECC组明显富集。Prevotella是辅助性T淋巴细胞(Th)17的强诱导剂,肠道Th17迁移可促进肿瘤发生发展[20-21],推测Prevotella诱导Th17的活化是ECC发生发展的一个重要环节。Vich等[22]的研究表明除抗生素外,抑酸药也会影响肠道菌群的多样性和物种结构。有研究[23]指出抑酸药可以通过提高胃肠道pH值抑制Prevotella,另外在一项研究[24]中发现抗生素治疗后患者肠道菌群的Bacteroidetes明显减少,同样本研究结果也显示抑酸药、抗生素与Bacteroidetes中的Prevotella呈负相关。本研究还发现Lactobacillus和Megasphaera在ECC组中明显增加,且与TBil、DBil等肝功能指标呈正相关,说明两者也与ECC疾病进展密切相关。Lactobacillus可以将初级胆汁酸转化为次级胆汁酸[25],报道[26-27]显示次级胆汁酸增多是胆管癌变的重要因素。ECC组Lactobacillus增加会代谢生成更多的次级胆汁酸,促进ECC的发生发展。Megasphaera是瘤胃微生物组的重要成员,正常情况下可生成短链脂肪酸,对肠道屏障功能具有保护作用[28]。Megasphaera扩增也可诱发炎症反应,从癌前病变至实性癌灶中均检测到Megasphaera。已经有研究者[29]提出Megasphaera在促进子宫宫颈发育异常和宫颈上皮内瘤变中起着重要作用,另有研究[30]发现与口腔癌患者相比,Megasphaera在口腔黏膜白斑患者中丰度也明显增加。Cortez等[31]发现在PSC中Megasphaera丰度增加,且Megasphaera与TBil、GGT呈正相关。而PSC是ECC常见的高危因素,本研究描述了Megasphaera在ECC组明显增多,这可能为进一步研究Megasphaera与ECC发生发展之间的关系提供一定的线索。Wu等[32]进一步发现Megasphaera可通过影响氨基酸代谢参与癌症疾病进展。
当然,本研究局限性在于样本数量较少,未来也会扩大样本量进一步研究探索肠道菌群变化与ECC的潜在联系。
综上所述,本研究表明,ECC患者肠道菌群发生明显改变,表现为Prevotella、Lactobacillus、Megasphaera、Sutterella明显增多,且ECC患者肠道菌群改变与肝功能和药物的使用密切相关。该研究将为从肠道菌群角度预防或治疗ECC提供研究基础。

-
表 1 3组一般资料比较
项目 ECC组(n=16) CBDS组(n=20) Normal组(n=10) 统计值 P值 男/女(例) 7/9 7/13 5/5 χ2=0.679 0.891 年龄(岁) 61.38±10.69 60.25±16.07 53.20±11.55 F=1.257 0.295 TBil(mol/L) 176.59(63.56~324.11) 29.24(14.74~113.73)1) 14.10(12.23~15.78)1) χ2=22.471 <0.001 DBil(mol/L) 150.16(55.11~278.45) 20.79(10.40~101.64)1) 6.96(6.18~8.35)1) χ2=24.575 <0.001 ALP(U/L) 268.65(200.25~437.00) 155.00(67.75~319.00)1) 77.50(68.25~89.00)1) χ2=18.447 <0.001 GGT(U/L) 379.00(226.50~774.75) 188.5(83.00~411.5)1) 33.00(21.00~39.50)1) χ2=23.311 <0.001 ALT(U/L) 80.50(48.25~102.98) 59.00(39.53~123.75) 21.50(18.75~29.00)1) χ2=17.767 <0.001 AST(U/L) 67.00(41.50~100.75) 46.00(27.25~85.00) 23.00(18.25~29.25)1) χ2=15.188 0.001 使用抑酸药天数(d) 13.00±10.67 16.54±8.25 t=0.290 0.885 使用保肝药天数(d) 12.00±5.38 6.60±9.11 t=0.843 0.724 使用抗生素天数(d) 12.33±18.06 14.77±2.34 t=0.621 0.967 PTCD治疗(例) 2 0 0.190 注: 与ECC组比较, 1)P<0.05。 表 2 3组间菌群多样性的比较
指数 ECC组(n=16) CBDS组(n=20) Normal组(n=10) F值 P值 Observed species 243±51 211±87 240±68 0.420 0.403 Chao1 244.43±51.59 213.24±88.55 240.98±67.62 0.420 0.421 Shannon 3.50±0.441) 2.48±0.99 3.37±0.381) 0.007 0.002 ACE 244.69±51.78 213.53±88.37 241.39±67.65 0.420 0.407 Simpson 0.072±0.0411) 0.259±0.210 0.091±0.1761) 0.002 <0.001 注: 与CBDS组比较,1)P<0.05。 -
[1] RIZVI S, KHAN SA, HALLEMEIER CL, et al. Cholangiocarcinoma-evolving concepts and therapeutic strategies[J]. Nat Rev Clin Oncol, 2018, 15(2): 95-111. DOI: 10.1038/nrclinonc.2017.157. [2] TYSON GL, EL-SERAG HB. Risk factors for cholangiocarcinoma[J]. Hepatology, 2011, 54(1): 173-184. DOI: 10.1002/hep.24351. [3] BIEDERMANN L, ROGLER G. The intestinal microbiota: Its role in health and disease[J]. Eur J Pediatr, 2015, 174(2): 151-167. DOI: 10.1007/s00431-014-2476-2. [4] AKSHINTALA VS, TALUKDAR R, SINGH VK, et al. The gut microbiome in pancreatic disease[J]. Clin Gastroenterol Hepatol, 2019, 17(2): 290-295. DOI: 10.1016/j.cgh.2018.08.045. [5] BIEDERMANN L, ROGLER G. The intestinal microbiota: Its role in health and disease[J]. Eur J Pediatr, 2015, 174(2): 151-167. DOI: 10.1007/s00431-014-2476-2. [6] HAN ML, GONG ZH. The mechanism of intestinal flora in hepatobiliary diseases[J]. J Clin Hepatol, 2017, 33(2): 384-388. DOI: 10.3969/j.issn.1001-5256.2017.02.040.韩美林, 龚振华. 肠道菌群在肝胆疾病中的作用机制[J]. 临床肝胆病杂志, 2017, 33(2): 384-388. DOI: 10.3969/j.issn.1001-5256.2017.02.040. [7] BENSON AB, D'ANGELICA MI, ABBOTT DE, et al. Hepatobiliary cancers, version 1.2020, NCCN clinical practice guidelines in oncology[J]. J Natl Compr Canc Netw, 2020, 10(5): 541-565. DOI: 10.6004/jnccn.2020.0036. [8] WILLIAMS E, BECKINGHAM I, EL SAYED G, et al. Updated guideline on the management of common bile duct stones (CBDS)[J]. Gut, 2017, 66(5): 765-782. DOI: 10.1136/gutjnl-2016-312317. [9] DOHERTY B, NAMBUDIRI VE, PALMER WC. Update on the diagnosis and treatment of cholangiocarcinoma[J]. Curr Gastroenterol Rep, 2017, 19(1): 2. DOI: 10.1007/s11894-017-0542-4. [10] JANG JY, KIM SW, PARK DJ, et al. Actual long-term outcome of extrahepatic bile duct cancer after surgical resection[J]. Ann Surg, 2005, 241(1): 77-84. DOI: 10.1097/01.sla.0000150166.94732.88. [11] LEE H, LEE HK, MIN SK, et al. 16S rDNA microbiome composition pattern analysis as a diagnostic biomarker for biliary tract cancer[J]. World J Surg Oncol, 2020, 18(1): 19. DOI: 10.1186/s12957-020-1793-3. [12] CHEN B, FU SW, LU L, et al. A Preliminary study of biliary microbiota in patients with bile duct stones or distal cholangiocarcinoma[J]. Biomed Res Int, 2019, 2019: 1092563. DOI: 10.1155/2019/1092563. [13] AVILÉS-JIMÉNEZ F, GUITRON A, SEGURA-LÓPEZ F, et al. Microbiota studies in the bile duct strongly suggest a role for Helicobacter pylori in extrahepatic cholangiocarcinoma[J]. Clin Microbiol Infect, 2016, 22(2): 178.e11-e22. DOI: 10.1016/j.cmi.2015.10.008. [14] WANG Q, HAO C, YAO W, et al. Intestinal flora imbalance affects bile acid metabolism and is associated with gallstone formation[J]. BMC Gastroenterol, 2020, 20(1): 59. DOI: 10.1186/s12876-020-01195-1. [15] WANG P, WANG Y, LU L, et al. Alterations in intestinal microbiota relate to intestinal failure-associated liver disease and central line infections[J]. J Pediatr Surg, 2017, 52(8): 1318-1326. DOI: 10.1016/j.jpedsurg.2017.04.020. [16] HⅡPPALA K, KAINULAINEN V, KALLIOMÄKI M, et al. Mucosal prevalence and interactions with the epithelium indicate commensalism of sutterella spp[J]. Front Microbiol, 2016, 7: 1706. DOI: 10.3389/fmicb.2016.01706. [17] KAAKOUSH NO. Sutterella species, iga-degrading bacteria in ulcerative colitis[J]. Trends Microbiol, 2020, 28(7): 519-522. DOI: 10.1016/j.tim.2020.02.018. [18] WEXLER HM. Bacteroides: The good, the bad, and the nitty-gritty[J]. Clin Microbiol Rev, 2007, 20(4): 593-621. DOI: 10.1128/CMR.00008-07. [19] GIBⅡNO G, LOPETUSO LR, SCALDAFERRI F, et al. Exploring bacteroidetes: Metabolic key points and immunological tricks of our gut commensals[J]. Dig Liver Dis, 2018, 50(7): 635-639. DOI: 10.1016/j.dld.2018.03.016. [20] YANG PJ, HUANG W, LIU HB. The correlation between IL-17 and liver disease[J]. J Clin Hepatol, 2017, 33(9): 1810-1814. DOI: 10.3969/j.issn.1001-5256.2017.09.041.杨浦娟, 黄祎, 刘华宝. IL-17与肝脏疾病的相关性[J]. 临床肝胆病杂志, 2017, 33(9): 1810-1814. DOI: 10.3969/j.issn.1001-5256.2017.09.041. [21] BELLONE M, BREVI A, HUBER S. Microbiota-propelled T helper 17 cells in inflammatory diseases and cancer[J]. Microbiol Mol Biol Rev, 2020, 84(2): e00064-19. DOI: 10.1128/MMBR.00064-19. [22] VICH VILA A, COLLIJ V, SANNA S, et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota[J]. Nat Commun, 2020, 11(1): 362. DOI: 10.1038/s41467-019-14177-z. [23] ZOU ZH, LIU D, LI HD, et al. Prenatal and postnatal antibiotic exposure influences the gut microbiota of preterm infants in neonatal intensive care units[J]. Ann Clin Microbiol Antimicrob, 2018, 17(1): 9. DOI: 10.1186/s12941-018-0264-y. [24] PLETZ MW, RAU M, BULITTA J, et al. Ertapenem pharmacokinetics and impact on intestinal microflora, in comparison to those of ceftriaxone, after multiple dosing in male and female volunteers[J]. Antimicrob Agents Chemother, 2004, 48(10): 3765-3772. DOI: 10.1128/AAC.48.10.3765-3772.2004. [25] ZENG H, LARSON KJ, CHENG WH, et al. Advanced liver steatosis accompanies an increase in hepatic inflammation, colonic, secondary bile acids and Lactobacillaceae/Lachnospiraceae bacteria in C57BL/6 mice fed a high-fat diet[J]. J Nutr Biochem, 2020, 78: 108336. DOI: 10.1016/j.jnutbio.2019.108336. [26] LONG Z, KONG D. Research progress on pathogenic factors of extrahepatic cholangiocarcinoma[J]. Jiangxi Trad Chin Med, 2017, 48(8): 72-75. https://www.cnki.com.cn/Article/CJFDTOTAL-JXZY201708028.htm龙祯, 孔棣. 肝外胆管癌致病因素的研究进展[J]. 江西中医药, 2017, 48(8): 72-75. https://www.cnki.com.cn/Article/CJFDTOTAL-JXZY201708028.htm [27] OHTANI N, KAWADA N. Role of the gut-liver axis in liver inflammation, fibrosis, and cancer: A special focus on the gut microbiota relationship[J]. Hepatol Commun, 2019, 3(4): 456-470. DOI: 10.1002/hep4.1331. [28] SHETTY SA, MARATHE NP, LANJEKAR V, et al. Comparative genome analysis of Megasphaera sp. reveals niche specialization and its potential role in the human gut[J]. PLoS One, 2013, 8(11): e79353. DOI: 10.1371/journal.pone.0079353. [29] AUDIRAC-CHALIFOUR A, TORRES-POVEDA K, BAHENA-ROMÁN M, et al. Cervical microbiome and cytokine profile at various stages of cervical cancer: A pilot study[J]. PLoS One, 2016, 11(4): e0153274. DOI: 10.1371/journal.pone.0153274. [30] GOPINATH D, KUNNATH MENON R, CHUN WIE C, et al. Salivary bacterial shifts in oral leukoplakia resemble the dysbiotic oral cancer bacteriome[J]. J Oral Microbiol, 2020, 13(1): 1857998. DOI: 10.1080/20002297.2020.1857998. [31] CORTEZ RV, MOREIRA LN, PADILHA M, et al. Gut microbiome of children and adolescents with primary sclerosing cholangitis in association with ulcerative colitis[J]. Front Immunol, 2020, 11: 598152. DOI: 10.3389/fimmu.2020.598152. [32] WU J, ZHANG C, XU S, et al. Fecal microbiome alteration may be a potential marker for gastric cancer[J]. Dis Markers, 2020, 2020: 3461315. DOI: 10.1155/2020/3461315. 期刊类型引用(2)
1. 俞雪,沈天皓,周诚,刘雨,李炜,蒋霆辉,朱永强,刘艳. 胆-肠轴在肝内胆管癌发生发展中的作用机制. 临床肝胆病杂志. 2025(03): 588-593 .  本站查看
本站查看2. 陈文辉,于龙,刘文韬,翟文龙. 基于高通量测序技术的胆囊癌患者肠道菌群变化特点. 河南医学研究. 2022(24): 4431-4437 .  百度学术
百度学术其他类型引用(0)
-




 PDF下载 ( 5925 KB)
PDF下载 ( 5925 KB)

 下载:
下载:





 下载:
下载:


 百度学术
百度学术