叉头转录因子A2、J2在肝细胞癌组织中的表达及意义
DOI: 10.3969/j.issn.1001-5256.2021.06.025
Expression and clinical significance of forkhead box A2 and forkhead box J2 in hepatocellular carcinoma
-
摘要:
目的 检测肝细胞癌(HCC)组织中叉头(FOX)转录因子A2(FOXA2)与FOXJ2的表达水平,探讨FOXA2、FOXJ2与HCC的相关性。 方法 选取2014年1月—2019年7月于河南中医药大学第一附属医院病理学检查明确为HCC患者54例,收集临床资料和病理组织。采用免疫组化S-P法检测HCC组织中FOXA2与FOXJ2的蛋白水平表达,分析其与HCC相关临床病理特征及患者预后的关系。计数资料比较采用χ2检验与校正χ2检验,FOXJ2与FOXA2表达相关性采用Spearman相关分析法;采用Kaplan-Merier法进行生存期分析;采用Image-Pro Plus进行FOXA2、FOXJ2表达的半定量分析;采用Wilcoxon秩和检验比较组间差异。 结果 HCC组织中FOXA2和FOXJ2蛋白表达阳性率分别为70.37%(38/54)和75.92%(41/54);FOXA2和FOXJ2表达水平间呈显著正相关(rs=0.648,P<0.001)。FOXA2阴性组与阳性组相比,肿瘤直径、分化程度、肿瘤个数、AFP水平差异均有统计学意义(χ2值分别为5.440、4.113、4.352、3.865,P值分别为0.020、0.043、0.037、0.049);FOXJ2阴性组与阳性组比较,分化程度差异有统计学意义(χ2=9.267,P=0.002)。FOXA2、FOXJ2阳性表达组HCC患者的累积生存率均显著高于FOXA2、FOXJ2阴性表达组HCC患者(P值均<0.01)。 结论 FOXA2与FOXJ2的表达水平可能与HCC的发生发展及预后相关,二者在HCC的发生发展中起协同作用。 Abstract:Objective To investigate the expression levels of forkhead box A2 (FOXA2) and forkhead box J2 (FOXJ2) in hepatocellular carcinoma (HCC) tissue and the association of FOXA2 and FOXJ2 with HCC. Methods Clinical data and pathological tissue samples were collected from 54 patients with pathologically confirmed HCC in The First Affiliated Hospital of Henan University of Traditional Chinese Medicine from January 2014 to July 2019. The immunohistochemical SP method was used to measure the protein expression levels of FOXA2 and FOXJ2 in HCC tissue, and their association with HCC-related clinicopathological features and patient prognosis was analyzed. The chi-square test and the adjusted chi-square test were used for comparison of categorical data; a Spearman correlation analysis was performed to investigate the correlation between the expression of FOXA2 and FOXJ2; the Kaplan-Meier method was used for survival analysis; Image-Pro Plus was used to perform the semi-quantitative analysis of the expression of FOXA2 and FOXJ2; the Wilcoxon rank-sum test was used for comparison between groups. Results The positive rates of the protein expression of FOXA2 and FOXJ2 in HCC tissue were 70.37% (38/54) and 75.92% (41/54), respectively, and there was a significant positive correlation between the expression levels of FOXA2 and FOXJ2 (rs=0.648, P < 0.001). In both negative and positive groups, the expression level of FOXA2 was associated with tumor diameter, degree of tumor differentiation, number of tumors, and alpha-fetoprotein (χ2=5.440, 4.113, 4.352, and 3.865, P=0.020, 0.043, 0.037, and 0.049), and the expression level of FOXJ2 was associated with the degree of tumor differentiation (χ2=9.267, P=0.002). The group with positive expression of FOXA2 and FOXJ2 had a significantly higher cumulative survival rate than the group with negative expression of FOXA2 and FOXJ2 (P < 0.01). Conclusion The expression levels of FOXA2 and FOXJ2 are associated with the development, progression, and prognosis of HCC, and they have a synergistic effect in the development and progression of HCC. -
Key words:
- Carcinoma, Hepatocellular /
- Forkhead Transcription Factors /
- Prognosis
-
原发性肝癌的发病率和病死率均较高,是目前我国第4位常见恶性肿瘤及第2位肿瘤致死病因[1-2],其中肝细胞癌(HCC)约占90%[3]。手术切除、介入、靶向药物等是目前肝癌治疗的常见手段[4-6],这些传统的治疗方法虽然能够取得一定疗效,但仍然不能有效降低肝癌病死率。因此,探讨肝癌发生发展的机制,筛选新的治疗方法及靶点,是目前肝癌研究的一个主要方向,对于提高肝癌患者总体预后具有重要的指导意义及临床应用价值。叉头(forkhead box,FOX)转录因子家族是进化上保守的转录因子超家族,其成员是参与转录调节和DNA修复的DNA结合蛋白,大多数成员与癌症有关,FOX家族基因通过基因扩增、逆转录病毒整合、染色体易位和转录调控参与致癌作用[7-9],因此,FOX转录因子可作为预测和治疗的直接或间接靶标,成为新型的监测标志物[10]。其中,FOXA2与FOXJ2是主要发挥抑癌作用的转录激活因子[11-12]。本研究通过免疫组化方法检测FOXA2、FOXJ2在HCC中的表达并探讨其临床意义。
1. 资料与方法
1.1 研究对象
选取2014年1月—2019年7月于河南中医药大学第一附属医院行手术或穿刺术的原发性肝癌患者54例,其中男44例,女10例,年龄29~78岁,中位年龄56岁,平均(54.85±1.52)岁。所有患者病理学检查证实为HCC,临床病理资料完整,均有乙型肝炎病史,未行放疗、化疗、生物治疗等相关治疗。参照《原发性肝癌诊疗规范(2019年版)》[1]进行分期,Ⅰ~Ⅱ期43例,Ⅲ~Ⅳ期11例。
1.2 试剂
免疫组化所需FOXA2抗体、DAB显色试剂盒均购自武汉博士德生物工程有限公司;FOXJ2抗体购自苏州百远生物科技有限公司;鼠/兔通用二抗(PV-9000)购自北京中杉金桥生物技术有限公司。
1.3 实验方法
石蜡组织切片需二甲苯脱蜡3次,每次10 min,梯度酒精水化,使用高压蒸汽锅进行高温抗原修复,室温下湿盒孵育,阻断内源性过氧化物酶孵育10 min,P BS冲洗3次,FOXA2、FOXJ2抗体均按照1∶ 100稀释,滴加一抗后37 ℃保温箱孵育60 min,PBS冲洗3次,滴加适量反应增强液,室温孵育20 min,PBS冲洗3次,室温孵育二抗20 min,PBS冲洗3次,加入新鲜配置的DAB显色液,室温孵育5 min,苏木素复染,脱水透明封片。
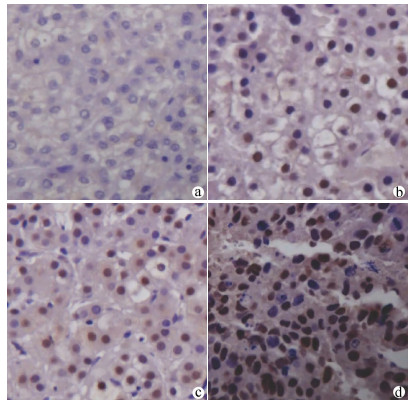
1.4 结果判定
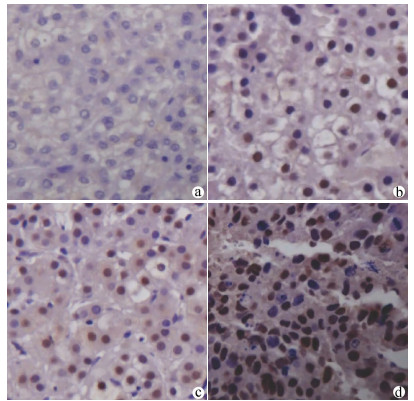
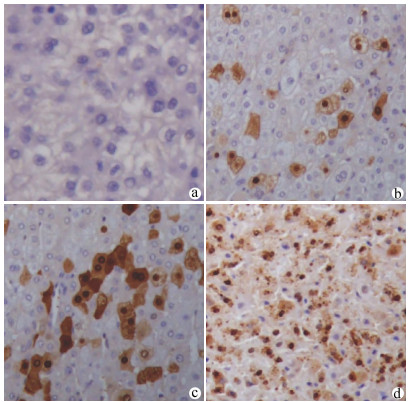
FOXA2、FOXJ2免疫组化染色以出现棕黄色或棕褐色为其阳性判定标准,FOXA2主要以细胞核染色为主,FOXJ2细胞核及胞质均染色。所有切片均由本院2名病理科医师采用双盲法进行观察,每张切片随机观察计数5个高倍镜视野下阳性细胞数,根据细胞的染色强度、细胞阳性率和半定量分析进行判定。高倍镜下随机选取10个高倍视野(×400),以阳性细胞所占的百分比联合细胞的染色强度进行评分。评分标准[13]如下:“-”不表达,计0分,“+”为低表达,计1 ~3分,“+ +”为中度表达,计4~6分,“+ + +”为高表达,计7~9分;0分为阴性,1~9分为阳性。
1.5 伦理学审查
本研究经由河南中医药大学第一附属医院伦理委员会审核,批号:2018HL-079-01。本研究中样本信息对外提供或开放使用已在中国人类遗传资源信息办公室备案,备案号:2021BAT0182。所有患者均签署知情同意书。
1.6 统计学方法
采用SPSS 21.0软件进行数据统计。计量资料用x ±s表示。肝组织中FOXA2、FOXJ2的表达与肝癌临床病理特征及AFP之间关系的计数资料比较采用χ2检验与校正χ2检验;FOXJ2与FOXA2表达相关性采用Spearman相关分析法;采用Kaplan-Merier法进行生存期分析;采用Image-Pro Plus进行FOXA2、FOXJ2表达的半定量分析;采用Wilcoxon秩和检验比较组间差异。P<0.05为差异有统计学意义。
2. 结果
2.1 FOXA2、FOXJ2在HCC组织中的表达及相关性分析
FOXA2在HCC组织呈弥漫性胞核表达,FOXJ2呈弥漫性胞核及细胞质表达,染色均呈棕黄色或棕褐色。54例HCC组织中,FOXA2阳性表达38例,阳性表达率约为70.37%(图 1);FOXJ2阳性表达41例,阳性表达率约为75.92%(图 2),二者半定量分析均值见表 1。HCC组织中FOXA2和FOXJ2表达水平均不服从正态分布,相关性分析采用Spearman秩相关分析,rs=0.648,P<0.001,提示FOXA2与FOXJ2表达水平之间呈显著正相关。
表 1 FOXA2、FOXJ2表达的半定量分析项目 FOXA2 FOXJ2 Area IOD Area IOD 不表达 24.60±13.35 3.51±1.91 24.60±13.35 2.96±1.63 低表达 204.53±147.05 84.15±70.3 175.46±537.81 13.14±110.56 中度表达 310.03±214.34 89.02±69.06 400.69±1 551.80 180.63±695.20 高表达 577.11±1212.88 445.26±939.64 1 505.42±9 001.71 652.10±3 987.02 2.2 HCC组织中FOXA2、FOXJ2的表达与临床病理特征的关系
FOXA2阳性组与阴性组间肿瘤直径、分化程度、肿瘤个数、AFP水平比较差异均有统计学意义(P值均<0.05);FOXJ2阳性组与阴性组间分化程度比较差异有统计学意义(P<0.05)(表 2)。
表 2 FOXA2、FOXJ2的表达与肝癌相关临床病理特征的关系项目 例数 FOXA2 χ2值 P值 FOXJ2 χ2值 P值 阴性(n=16) 阳性(n=38) 阴性(n=13) 阳性(n=41) 性别(例) 0 1.000 0.006 0.940 男 44 13 31 10 34 女 10 3 7 3 7 年龄(例) 0.558 0.455 0.001 0.971 <60岁 33 11 22 8 25 ≥60岁 21 5 16 5 16 肿瘤直径(例) 5.440 0.020 2.026 0.155 ≤5 cm 30 5 25 5 25 >5 cm 24 11 13 8 16 分化程度(例) 4.113 0.043 9.267 0.002 低/中分化 11 6 5 7 4 高分化 43 10 33 6 37 TNM分期(例) 0.032 0.859 0.453 0.501 Ⅰ~Ⅱ期 43 12 31 9 34 Ⅲ~Ⅳ期 11 4 7 4 7 门静脉癌栓(例) 1.072 0.300 2.612 0.106 有 12 5 7 5 7 无 42 11 31 8 34 转移(例) 0.012 0.913 0 1.000 有 8 3 5 2 6 无 46 13 33 11 35 肿瘤个数(例) 4.352 0.037 1.621 0.203 单个 27 4 23 4 23 多个 27 12 15 9 18 AFP(例) 3.865 0.049 2.038 0.153 ≤20 ng/ml 28 5 23 4 24 >20 ng/ml 26 11 15 9 17 采用Wilcoxon秩和检验对与FOXA2和FOXJ2表达水平相关的病理特征组间差异进行比较,结果提示,FOXA2:肿瘤直径大小(Z=-0.518,P=0.604)、AFP表达水平高低(Z=-1.386,P=0.166)组间比较差异均无统计学意义,分化程度高低(Z=-2.413,P=0.016)、肿瘤个数多少(Z=-2.113,P=0.035)组间比较差异均有统计学意义;FOXJ2:分化程度高低(Z=-3.423,P=0.001)组间比较差异有统计学意义。
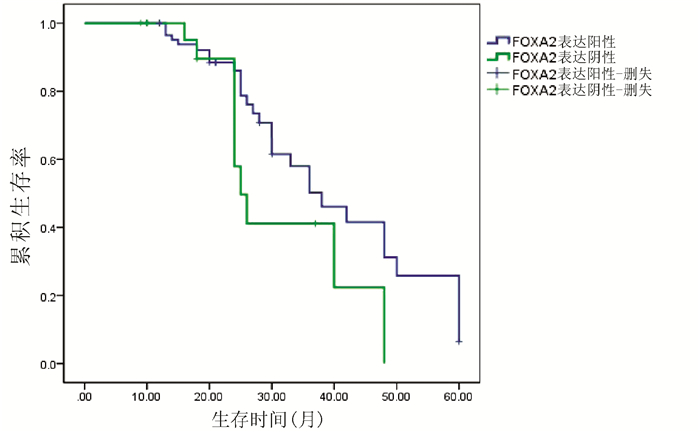
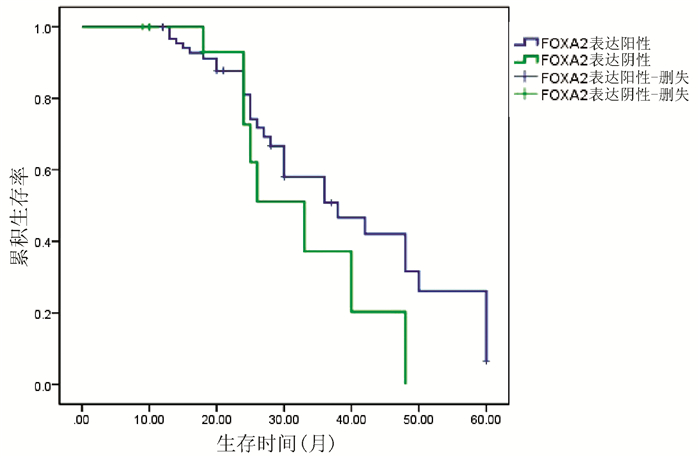
2.3 HCC组织中FOXA2、FOXJ2的表达与预后的关系
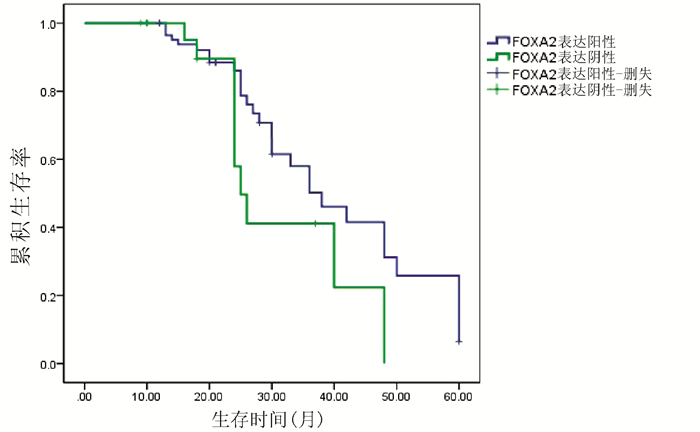
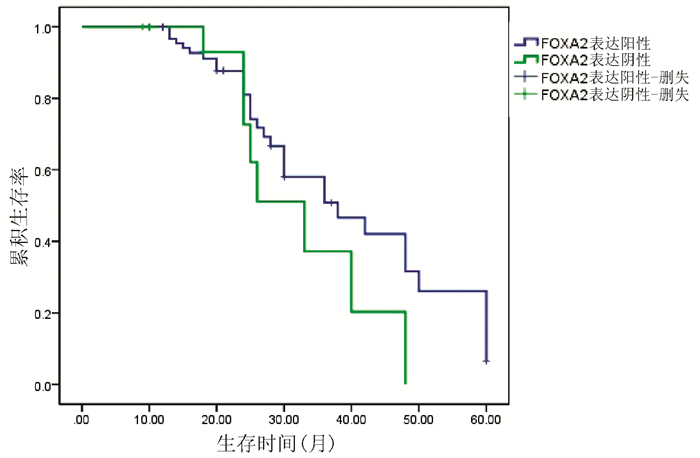
对54例HCC患者进行Kaplan-Meier生存曲线分析,结果显示,FOXA2、FOXJ2阳性表达组HCC患者的累积生存率均显著高于FOXA2、FOXJ2阴性表达组HCC患者(P值均<0.01)(图 3、4)。
3. 讨论
HCC治疗后复发率高,转移率高,预后差,尽管新型药品及医疗技术的进步使患者生存状况较前改善,但仍然无法满足治疗需要[4-6]。目前癌症的研究主要集中在人类癌症分子靶向相关治疗策略的开发上[14]。过去10年中,分子细胞生物学的进展进一步阐明了癌症生长、侵袭和转移的机制,并促进了分子靶向药物的开发[15]。转录调控是基因调控最常见的方式之一,转录因子通过结合特定的DNA序列来控制从DNA到信使RNA的遗传信息转录速率,转录多种因素包括引发和调节基因转录的多种蛋白质[16-17]。FOX家族是进化上保守的转录因子超家族,其特点在于独特的DNA结合叉头结构域,FOX结构域是一个约有100个氨基酸的单体DNA结合结构域,在生物过程中起着调节细胞生长、分化、代谢、增殖、凋亡、迁移、侵袭以及胚胎发生、发育和寿命等多种作用[9, 18]。FOX家族基因作为癌基因或肿瘤抑制基因参与癌症的发生及发展[7-8],不同的亚家族其作用机制不同[19]。人类癌症的全基因组测序数据的快速发展,将揭开FOX家族基因本身及其FOX结合位点在肿瘤细胞上的突变情况,最终将应用于临床上癌症的诊断、治疗及预后评估[20]。
FOXA2又被称为肝细胞核因子3-β,是与肝特异性白蛋白相关的转录激活因子,FOXA2与染色质相互作用,通过调控基因转录和翻译,参与机体的生长发育及能量代谢[21-23]。同时,FOXA2也是肝脏发育的先驱因子,具有调节肝脏发育和代谢平衡的作用[24-25]。近年来,有多篇文献[26-30]报道FOXA2具有抑癌作用,在前列腺癌、子宫内膜癌、肺癌、胃癌、神经胶质瘤等肿瘤细胞中均呈下调趋势,其表达水平与癌症恶性程度呈负相关。Wang等[31]对比肝癌组织与癌旁组织中FOXA2表达,发现FOXA2在癌组织中的表达明显降低,FOXA2通过抑制基质金属蛋白酶-9的转录和表达,进而抑制HCC的侵袭性和转移,并影响了上皮细胞-间充质转化相关蛋白的表达水平。FOXA2的高表达抑制了肝癌细胞增殖,迁移和侵袭的能力,FOXA2可能成为治疗HCC的新靶点[32]。本研究结果显示,FOXA2的表达水平与TNM分期、癌栓形成、是否转移均无关,这一结论有待扩大样本量进一步研究;FOXA2的表达水平与肿瘤直径、分化程度、肿瘤个数、AFP相关,并且与分化程度高低、肿瘤个数多少相关;FOXA2阳性表达组HCC患者的生存时间显著高于FOXA2阴性表达组,提示FOXA2的表达水平与HCC的发生发展及预后相关,可能是影响肝癌预后的重要基因。
FOXJ2是双序列特异性结合DNA,有FHX.L和FHX.S(由位于人类染色体上的单个基因编码)2种同工型[33],其重要性最近被广泛关注[34]。FOXJ2目前定位于细胞核中,并且可能参与所有FOX因子的核易位机制[35],在参与人类早期胚胎发育等诸多过程中发挥重要作用,与调控细胞的生长及代谢以及细胞多种生物学特性关系密切[36]。研究[13, 37-39]证实,FOXJ2可抑制鼻咽癌、脑胶质瘤、肺癌、乳腺癌等肿瘤细胞的侵袭和转移,其表达水平在肿瘤组织中下调,且表达水平降低与肿瘤患者预后不良呈正相关。FOXJ2也可能是肝癌预后的重要因素,Zhang等[40]通过免疫印迹法和免疫组织化学分析表明,FOXJ2在肝癌组织及肝癌细胞中均下调,并且观察到FOXJ2表达水平与肝癌之间呈负相关,并使用CCK-8分析法证实FOXJ2能够有效抑制HCC的增殖。本研究结果显示,FOXJ2的表达水平与HCC的分化程度及分化程度高低相关;FOXJ2阴性表达组HCC患者的生存时间显著低于FOXJ2阳性表达组患者。由此可见,FOXJ2的表达水平与HCC的发生发展及预后关系密切,表明FOXJ2可能是影响HCC患者预后的重要因素。本研究未能发现其与TNM分期、癌栓形成、是否远处转移的相关性,可能与本研究样本量、临床穿刺及手术患者分期较早等临床特点相关,有待进一步临床、基础研究探讨。
靶基因联合是目前备受关注的热点,联合治疗可大幅度提高患者获益率。FOXA2、FOXJ2作为FOX转录因子家族成员,通过发挥抑癌作用参与癌症的发生及发展。本研究结合临床数据,重点分析FOXA2、FOXJ2表达水平与临床相关临床病理特征及预后的关系,为肝癌治疗提供新思路。本研究临床数据证实,FOXA2和FOXJ2在HCC组织内的表达水平之间呈显著正相关,表明二者对HCC的影响起协同作用,未来有望共同参与HCC的预测及治疗。
-
表 1 FOXA2、FOXJ2表达的半定量分析
项目 FOXA2 FOXJ2 Area IOD Area IOD 不表达 24.60±13.35 3.51±1.91 24.60±13.35 2.96±1.63 低表达 204.53±147.05 84.15±70.3 175.46±537.81 13.14±110.56 中度表达 310.03±214.34 89.02±69.06 400.69±1 551.80 180.63±695.20 高表达 577.11±1212.88 445.26±939.64 1 505.42±9 001.71 652.10±3 987.02 表 2 FOXA2、FOXJ2的表达与肝癌相关临床病理特征的关系
项目 例数 FOXA2 χ2值 P值 FOXJ2 χ2值 P值 阴性(n=16) 阳性(n=38) 阴性(n=13) 阳性(n=41) 性别(例) 0 1.000 0.006 0.940 男 44 13 31 10 34 女 10 3 7 3 7 年龄(例) 0.558 0.455 0.001 0.971 <60岁 33 11 22 8 25 ≥60岁 21 5 16 5 16 肿瘤直径(例) 5.440 0.020 2.026 0.155 ≤5 cm 30 5 25 5 25 >5 cm 24 11 13 8 16 分化程度(例) 4.113 0.043 9.267 0.002 低/中分化 11 6 5 7 4 高分化 43 10 33 6 37 TNM分期(例) 0.032 0.859 0.453 0.501 Ⅰ~Ⅱ期 43 12 31 9 34 Ⅲ~Ⅳ期 11 4 7 4 7 门静脉癌栓(例) 1.072 0.300 2.612 0.106 有 12 5 7 5 7 无 42 11 31 8 34 转移(例) 0.012 0.913 0 1.000 有 8 3 5 2 6 无 46 13 33 11 35 肿瘤个数(例) 4.352 0.037 1.621 0.203 单个 27 4 23 4 23 多个 27 12 15 9 18 AFP(例) 3.865 0.049 2.038 0.153 ≤20 ng/ml 28 5 23 4 24 >20 ng/ml 26 11 15 9 17 -
[1] Bureau of Medical AdministrationNational Health Commission of the People's Republic of China. Guidelines for diagnosis and treatment of primary liver cancer in China (2019 edition)[J]. J Clin Hepatol, 2020, 36(2): 277-292. DOI: 10.3969/j.issn.1001-5256.2020.02.007.中华人民共和国国家卫生健康委员会医政医管局. 原发性肝癌诊疗规范(2019年版)[J]. 临床肝胆病杂志, 2020, 36(2): 277-292. DOI: 10.3969/j.issn.1001-5256.2020.02.007. [2] CHEN W, ZHENG R, BAADE PD, et al. Cancer statistics in China, 2015[J]. CA Cancer J Clin. 2016, 66(2): 115-32. DOI: 10.3322/caac.21338. [3] SIA D, VILLANUEVA A, FRIEDMAN SL, et al. Liver cancer cell of origin, molecular class, and effects on patient prognosis[J]. Gastroenterology, 2017, 152(4): 745-761. DOI: 10.1053/j.gastro.2016.11.048. [4] HAN JH, KIM DG, NA GH, et al. Evaluation of prognostic factors on recurrence after curative resections for hepatocellular carcinoma[J]. World J Gastroenterol, 2014, 20(45): 17132-17140. DOI: 10.3748/wjg.v20.i45.17132. [5] BOULIN M, DELHOM E, PIERREDON-FOULONGNE MA, et al. Transarterial chemoembolization for hepatocellular carcinoma: An old method, now flavor of the day[J]. Diagn Interv Imaging, 2015, 96(6): 607-615. DOI: 10.1016/j.diii.2015.04.005. [6] YANG J, YAN L, WANG W. Current status of multimodal & combination therapy for hepatocellular carcinoma[J]. Indian J Med Res, 2012, 136(3): 391-403. [7] WANG J, LI W, ZHAO Y, et al. Members of FOX family could be drug targets of cancers[J]. Pharmacol Ther, 2018, 181: 183-196. DOI: 10.1016/j.pharmthera.2017.08.003. [8] KATOH M, IGARASHI M, FUKUDA H, et al. Cancer genetics and genomics of human FOX family genes[J]. Cancer Lett, 2013, 328(2): 198-206. DOI: 10.1016/j.canlet.2012.09.017. [9] KATOH M, KATOH M. Human FOX gene family (Review)[J]. Int J Oncol, 2004, 25(5): 1495-1500. http://europepmc.org/abstract/MED/15492844 [10] MYATT SS, LAM EW. The emerging roles of forkhead box (Fox) proteins in cancer[J]. Nat Rev Cancer, 2007, 7(11): 847-859. DOI: 10.1038/nrc2223. [11] LIU J, YU Z, XIAO Y, et al. Coordination of FOXA2 and SIRT6 suppresses the hepatocellular carcinoma progression through ZEB2 inhibition[J]. Cancer Manag Res, 2018, 10: 391-402. DOI: 10.2147/CMAR.S150552. [12] ZHANG H, TANG QF, SUN MY, et al. ARHGAP9 suppresses the migration and invasion of hepatocellular carcinoma cells through up-regulating FOXJ2/E-cadherin[J]. Cell Death Dis, 2018, 9(9): 916. DOI: 10.1038/s41419-018-0976-0. [13] SHAN Y, CHANG T, SHI S, et al. Foxj2 overexpression is associated with poor prognosis, progression, and metastasis in nasopharyngeal carcinoma[J]. Onco Targets Ther, 2017, 10: 3733-3741. DOI: 10.2147/OTT.S134915. [14] TSUCHIYA N, SAWADA Y, ENDO I, et al. Biomarkers for the early diagnosis of hepatocellular carcinoma[J]. World J Gastroenterol, 2015, 21(37): 10573-10583. DOI: 10.3748/wjg.v21.i37.10573. [15] LI H, CHEN D, ZHANG J. Analysis of intron sequence features associated with transcriptional regulation in human genes[J]. PLoS One, 2012, 7(10): e46784. DOI: 10.1371/journal.pone.0046784. [16] LEE TI, YOUNG RA. Transcriptional regulation and its misregulation in disease[J]. Cell, 2013, 152(6): 1237-1251. DOI: 10.1016/j.cell.2013.02.014. [17] LEHMANN OJ, SOWDEN JC, CARLSSON P, et al. Fox's in development and disease[J]. Trends Genet, 2003, 19(6): 339-344. DOI: 10.1016/S0168-9525(03)00111-2. [18] HANNENHALLI S, KAESTNER KH. The evolution of Fox genes and their role in development and disease[J]. Nat Rev Genet, 2009, 10(4): 233-240. DOI: 10.1038/nrg2523. [19] JIN Y, LIANG Z, LOU H. The emerging roles of fox family transcription factors in chromosome replication, organization, and genome stability[J]. Cells, 2020, 9(1): 258. DOI: 10.3390/cells9010258. [20] BENAYOUN BA, CABURET S, VEITIA RA. Forkhead transcription factors: Key players in health and disease[J]. Trends Genet, 2011, 27(6): 224-232. DOI: 10.1016/j.tig.2011.03.003. [21] LI J, DANTAS MACHADO AC, GUO M, et al. Structure of the forkhead domain of FOXA2 bound to a complete DNA consensus site[J]. Biochemistry, 2017, 56(29): 3745-3753. DOI: 10.1021/acs.biochem.7b00211. [22] LIU J, YU Z, XIAO Y, et al. Coordination of FOXA2 and SIRT6 suppresses the hepatocellular carcinoma progression through ZEB2 inhibition[J]. Cancer Manag Res, 2018, 10: 391-402. DOI: 10.2147/CMAR.S150552. [23] LI Z, TUTEJA G, SCHUG J, et al. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer[J]. Cell, 2012, 148(1-2): 72-83. DOI: 10.1016/j.cell.2011.11.026. [24] WANG W, YAO LJ, SHEN W, et al. FOXA2 alleviates CCl4-induced liver fibrosis by protecting hepatocytes in mice[J]. Sci Rep, 2017, 7(1): 15532. DOI: 10.1038/s41598-017-15831-6. [25] YAMAMURA N, FUGO K, KISHIMOTO T. Forkhead box protein A2, a pioneer factor for hepatogenesis, is involved in the expression of hepatic phenotype of alpha-fetoprotein-producing adenocarcinoma[J]. Pathol Res Pract, 2017, 213(9): 1082-1088. DOI: 10.1016/j.prp.2017.07.024. [26] QI J, NAKAYAMA K, CARDIFF RD, et al. Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors[J]. Cancer Cell, 2010, 18(1): 23-38. DOI: 10.1016/j.ccr.2010.05.024. [27] SMITH B, NEFF R, COHN DE, et al. The mutational spectrum of FOXA2 in endometrioid endometrial cancer points to a tumor suppressor role[J]. Gynecol Oncol, 2016, 143(2): 398-405. DOI: 10.1016/j.ygyno.2016.08.237. [28] BASSERES DS, D'ALÒ F, YEAP BY, et al. Frequent downregulation of the transcription factor Foxa2 in lung cancer through epigenetic silencing[J]. Lung Cancer, 2012, 77(1): 31-37. DOI: 10.1016/j.lungcan.2012.01.011. [29] LI C, LU S, SHI Y. MicroRNA-187 promotes growth and metastasis of gastric cancer by inhibiting FOXA2[J]. Oncol Rep, 2017, 37(3): 1747-1755. DOI: 10.3892/or.2017.5370. [30] DING B, LIANG H, GAO M, et al. Forkhead Box A2 (FOXA2) inhibits invasion and tumorigenesis in glioma cells[J]. Oncol Res, 2017, 25(5): 701-708. DOI: 10.3727/096504016X14772378087005. [31] WANG J, ZHU CP, HU PF, et al. FOXA2 suppresses the metastasis of hepatocellular carcinoma partially through matrix metalloproteinase-9 inhibition[J]. Carcinogenesis, 2014, 35(11): 2576-2583. DOI: 10.1093/carcin/bgu180. [32] LIU J, YU Z, XIAO Y, et al. Coordination of FOXA2 and SIRT6 suppresses the hepatocellular carcinoma progression through ZEB2 inhibition[J]. Cancer Manag Res, 2018, 10: 391-402. DOI: 10.2147/CMAR.S150552. [33] PÉREZ-SÁNCHEZ C, ARIAS-DE-LA-FUENTE C, GÓMEZ-FERRERíA MA, et al. FHX. L and FHX. S, two isoforms of the human fork-head factor FHX (FOXJ2) with differential activity[J]. J Mol Biol, 2000, 301(4): 795-806. DOI: 10.1006/jmbi.2000.3999. [34] GRANADINO B, ARIAS-DE-LA-FUENTE C, PÉREZ-SÁNCHEZ C, et al. Fhx (Foxj2) expression is activated during spermatogenesis and very early in embryonic development[J]. Mech Dev, 2000, 97(1-2): 157-160. DOI: 10.1016/s0925-4773(00)00410-x. [35] GÓMEZ-FERRERÍA MA, REY-CAMPOS J. Functional domains of FOXJ2[J]. J Mol Biol, 2003, 329(4): 631-644. DOI: 10.1016/s0022-2836(03)00524-2. [36] MARTÍN-DE-LARA F, SÁNCHEZ-APARICIO P, ARIAS DE LA FUENTE C, et al. Biological effects of FoxJ2 over-expression[J]. Transgenic Res, 2008, 17(6): 1131-1141. DOI: 10.1007/s11248-008-9214-3. [37] QIU X, JI B, YANG L, et al. The role of FoxJ2 in the migration of human glioma cells[J]. Pathol Res Pract, 2015, 211(5): 389-397. DOI: 10.1016/j.prp.2015.01.005. [38] YANG Q, CAO X, TAO G, et al. Effects of FOXJ2 on TGF-β1-induced epithelial-mesenchymal transition through Notch signaling pathway in non-small lung cancer[J]. Cell Biol Int, 2017, 41(1): 79-83. DOI: 10.1002/cbin.10680. [39] WANG Y, YANG S, NI Q, et al. Overexpression of forkhead box J2 can decrease the migration of breast cancer cells[J]. J Cell Biochem, 2012, 113(8): 2729-2737. DOI: 10.1002/jcb.24146. [40] ZHANG Z, MENG G, WANG L, et al. The prognostic role and reduced expression of FOXJ2 in human hepatocellular carcinoma[J]. Mol Med Rep, 2016, 14(1): 254-262. DOI: 10.3892/mmr.2016.5261. 期刊类型引用(1)
1. 张鑫,冉小柯,陈茜,徐宗瑶,李艳杰,李闪宜,高琪,陈晓琦. 肝细胞癌患者血清中FOXA2与FOXJ2的表达及临床意义. 现代肿瘤医学. 2023(20): 3817-3821 .  百度学术
百度学术其他类型引用(0)
-




 PDF下载 ( 3112 KB)
PDF下载 ( 3112 KB)


 下载:
下载:



 下载:
下载:

 百度学术
百度学术