肝细胞癌分子异质性与临床精准治疗
DOI: 10.3969/j.issn.1001-5256.2021.08.006
Molecular heterogeneity of hepatocellular carcinoma and its precise treatment in clinical practice
-
摘要: 肝细胞癌是我国最常见的肝癌类型,分子异质性显著,极大地制约了临床疗效,因此深入探索其异质性的分子分型,对于制订个体化诊疗策略极为重要。近年来多种高通量测序技术层出不穷,结合多组学特征提出了多种分子分型系统,使临床研究者对肝癌分子异质性有了更加深刻的认识。详细总结了肝细胞癌的分子分型,探讨其与临床病理特征的密切联系,分析分子靶向治疗和免疫治疗的干预新靶标,提出了肝癌精准诊断和个体化治疗的新思路。Abstract: Hepatocellular carcinoma (HCC) is the most common type of liver cancer in China, and high molecular heterogeneity of HCC has caused the limitations in clinical efficacy; therefore, it is of great importance to explore its heterogeneity based on molecular classification and develop individualized diagnosis and treatment strategies. Various high-throughput sequencing techniques and multi-omics techniques in recent years have helped to establish multiple molecular classification systems, which gives us a deeper understanding of the molecular heterogeneity of HCC. This article summarizes the molecular classifications of HCC and discusses their association with clinicopathological features. This article also analyzes the new targets for molecular targeted therapy and immunotherapy and proposes new ideas for precise diagnosis and individualized treatment of HCC.
-
Key words:
- Carcinoma, Hepatocellular /
- Genetic Heterogeneity /
- Therapeutics
-
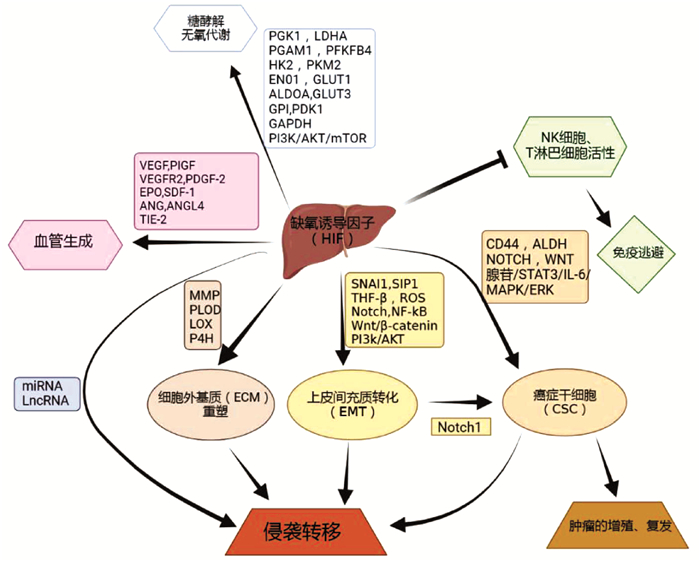
根世界卫生组织[1]估计,2030年将有100多万患者死于原发性肝癌。原发性肝癌包括肝细胞癌(HCC)、肝内胆管癌和混合型肝细胞癌-胆管癌,其中HCC占比约85%[2]。HCC的发生是一个逐步的过程。慢性肝炎病毒感染、过量饮酒和非酒精性脂肪性肝炎可导致肝脏炎症和组织损伤,是HCC的主要病因。肝损伤可扰乱肝血管系统、正常血流和O2供应,形成一个缺氧的微环境。缺氧的Kupffer细胞、巨噬细胞和肝细胞可激活肝脏中的肝星状细胞,加快胶原蛋白沉积,导致纤维化和肝硬化,从而进一步加剧缺氧。缺氧也可影响不同免疫细胞(如NK细胞)的活动,并抑制HCC微环境中许多不同类型免疫细胞的浸润和积累,包括肿瘤相关巨噬细胞(TAM)、髓源性抑制细胞(MDSC)和中性粒细胞。O2水平随着HCC的发展而降低,从而促进免疫抑制微环境的形成[3]。缺氧诱导因子(HIF)是参与细胞对缺氧适应的核心参与者,并受氧感应脯氨酰羟化酶的调节。缺氧通过氧化还原效应和HIF的稳定作用影响细胞生长的诸多方面。HIF亚型可能通过改变代谢、生长和自我更新对肿瘤生长产生不同的影响,并可能高度依赖于环境。HIF通过促进血管生成影响癌细胞的迁移、侵袭和转移、增殖,促进糖酵解,治疗抵抗,免疫逃避等方式影响HCC的发病和进展[4]。探究HIF对HCC的作用机制及临床意义,有望拓展诊治HCC的新方法。
1. HIF的特性
HIF是异二聚体,由α-亚基(HIF-α,包括HIF-1α,HIF-2α/EPAS1和HIF- 3α)和β-亚基(HIF-β,包括HIF-1β/ARNT1、ARNT2和ARNT3)组成。在人类和其他脊椎动物中,存在3种不同的HIF基因。在有氧(常氧)的情况下,HIF-α相互作用并与vonHippel-Lindau(VHL)蛋白结合,从而激活泛素连接酶系统,导致HIFa的蛋白酶体降解,HIF-1α蛋白的表达被抑制到非常低的水平。而在缺氧期间,脯氨酰羟化酶不活跃,导致HIF-α稳定并与HIF-1β二聚。二聚化后,HIF易位至细胞核,与包含序列5[0]-[A/G]CGTG-3[0]的启动子区域内的E-box样缺氧反应元件结合。HIF激活控制细胞氧稳态的基因,包括红细胞生成/血管生成和线粒体代谢有关的基因。缺氧细胞通过主要由HIF调控的转录和转录后机制对抗压力。这些分子变化使细胞能够适应缺氧,主要通过转用糖酵解来降低耗氧量,并减少细胞分裂(如细胞分裂)所需的能量。大多数实体瘤都有一定程度的缺氧,这与临床疗效有关。HIF活性的诱导上调了涉及癌症许多特征的基因,包括代谢重编程,细胞增殖、侵袭和转移以及对治疗的抵抗[5-6]。
除了氧依赖性机制外,HIF的表达和活性还受氧非依赖性机制的控制,这些机制调节了基因转录、mRNA翻译、蛋白质-蛋白质相互作用和HIF-1α亚基的翻译后修饰。在炎症反应中,HIF-1α基因的转录上调是以NF-κB依赖性方式实现的,并涉及信号转导和转录激活因子3(signal transducers and activators of transcription 3,STAT3)和Sp1。此外,生长因子对PI3K/ AKT通路的激活可导致HIF-1α mRNA和蛋白质合成增加。HIF-1α也通过与其他蛋白质的结合而受到调节[7]。
2. HIF在HCC中的作用机制
2.1 HIF通过促进糖酵解使HCC适应低氧应激
常氧环境中的细胞将葡萄糖转化为丙酮酸,丙酮酸进入三羧酸循环并在线粒体中氧化磷酸化,最终产生三磷酸腺苷。肿瘤细胞则表现出葡萄糖消耗增加和向糖酵解的重要代谢转变,丙酮酸转化为乳酸。即使在有氧条件下,肿瘤细胞中的这种代谢转变依然存在,被称为Warburg效应[8]。通过AKT和HIF-1α介导的MAP17表达上调,将促进Warburg效应[9]。肿瘤细胞通过在无氧糖酵解中来产生能量,相关研究发现这一过程主要由HIF-1α调节。许多参与糖酵解的关键酶是HCC细胞中直接的HIF-1α靶标,包括PGK1、PGAM1、HK2、ENO1、ALDOA、GPI、GAPDH、LDHA、PFKFB4和PKM2等[10],同时HIF-1α可诱导多种糖酵解蛋白同种型(包括GLUT1和GLUT3)的过度表达和活性增加,这对于糖酵解通量控制十分重要[11]。此外,HIF-1α增加了线粒体相关酶的表达,如丙酮酸脱氢酶激酶1,其可抑制丙酮酸转化为乙酰辅酶A,从而减少线粒体的氧化磷酸化水平和耗氧量[12]。而HIF-2α影响脂质代谢来进行能量代谢,如缺氧下,PI3K/AKT/mTOR通路可能是HIF-2α调控HCC的脂质代谢途径,进而促进HCC适应无氧环境的重要机制[13]。总的来说,研究显示HIF从不同途径促进了HCC的糖酵解,以适应低氧应激。
2.2 HIF通过促进血管生成促进HCC生长
HIF-1α是血管内皮生长因子(VEGF)表达的主要调节因子。在缺氧条件下,高水平积累的HIF-1α上调VEGF等一系列血管生成因子的表达,增强相关mRNA的转录,最后促进肿瘤血管生成[14]。HIF-2α也是调节肝细胞中VEGF和其他血管生成因子的主要HIF[15]。HIF-2α主要作用于血管生成相关基因,包括VEGF、促红细胞生成素、VEGF受体2(VEGFR2)、血管生成素和酪氨酸蛋白激酶受体TIE-2[16]。
除VEGF外,许多其他信号分子在缺氧条件下也通过HIF依赖性机制高表达,包括胎盘生长因子、血小板衍生生长因子β和基质衍生因子1[17-19],均可促进肿瘤中的血管生成。ANG样蛋白4也被鉴定为HIF-1α的基因靶标,其可通过调节血管细胞黏附分子和整合素β1的表达来影响HCC血管生成和转移[20]。因此,HIF促瘤血管的生成可促进HCC的生长。
2.3 HIF通过介导侵袭转移促进HCC进展
HIF诱导的肿瘤扩散的机制包括上皮间充质转化(epithelial-mesenchymal transition,EMT)、TWIST等基因的激活和GLUT1等参与癌症代谢的转录因子的激活。EMT涉及多种类型的肿瘤和多种肿瘤转移机制[21]。在HCC细胞中,缺氧条件下可通过SNAI1、SIP1、TGFβ、ROS、Notch、NF-κB、Wnt/β-catenin、PI3K/AKT等途径的活化来诱导EMT,上皮细胞转变为可移动的基质细胞,获得迁移到远处部位的能力[22]。细胞外基质(ECM)重塑在肿瘤侵袭和转移中发挥着至关重要的作用。参与ECM的几种酶沉积和重塑受缺氧和HIF的调节,包括基质金属蛋白酶、2-酮戊二酸5- 双加氧酶、赖氨酰氧化酶、胶原蛋白脯氨酰4-羟化酶等,同时通过HIF-1α依赖性机制,可下调HCC细胞中金属蛋白酶2组织抑制剂的表达[23]。HIF-1α/LOX通路通过依赖于肝炎反式激活蛋白X的机制,参与ECM重塑和促进HCC转移[24]。
此外,微小RNA(microRNA,miRNA)也与肿瘤的迁移和转移密切相关。例如,miR-23是一种常见的致癌基因,研究[25]显示,miR-23a/b可以通过VHL-HIF-1α通路促进HCC的发病进展。通过抑制SMC4可显著抑制HCC细胞的迁移,而HIF-1可通过转录调节和抑制miR-219来增加SMC4的表达。因此,HIF-1/miR-219/SMC4调控通路可能是促进HCC的迁移和转移一种机制[26]。此外,HIF-1α/miR-671-5p/TUFT1/AKT也可能是一种机制[27]。lncRNA也可促进HCC转移,lncRNA锌指蛋白多型2反义RNA1通过调控miR-576-3p/HIF-1α轴促进HCC的增殖和迁移[28]。此外,lncRNA可抑制HCC的转移,如lincRNA-p21可下调HIF-1α来降低VEGF水平,从而抑制HCC的侵袭能力[29]。因此,需要更多研究从非编码RNA层面来阐明HIF的作用机制,挖掘非编码RNA靶向药物。
2.4 HIF通过介导免疫逃避、癌症干细胞促进HCC发生发展
缺氧与HIF和肿瘤细胞逃避免疫反应有关。免疫细胞的功能受HIF1依赖性信号机制的调节,在缺氧期间,HIF诱导肿瘤细胞对CD8细胞毒性T淋巴细胞和NK细胞产生抗性,其可能是HIF通过AMPK/CREB信号增加IL-10表达,分泌的IL-10通过STAT3信号通路抑制NK细胞的细胞毒性,从而促进HCC的复发和转移[30]。此外,缺氧可以上调产生腺苷的细胞外酶CD39和CD73的表达,增加其在细胞环境中的浓度。腺苷通过与其A2A受体结合,强烈抑制活化的T淋巴细胞和NK细胞的抗肿瘤功能[31]。HIF还可以通过作用于TAM来抑制对肿瘤的免疫反应。研究[32]认为,TAM通过分泌细胞因子,如IL-10、TGFβ、IL-6、VEGF和IL-8,促进肿瘤细胞的生长、侵袭和转移。
MDSC具有免疫抑制活性,可使癌症逃避免疫监视并对免疫无反应。通过MDSC介导L-精氨酸的消耗,阻碍T淋巴细胞增殖,并与T淋巴细胞受体亚基CD3的下调相关,导致T淋巴细胞受体反应降低。肝肿瘤的MDSC水平升高,MyD88-NF-κB通路的激活刺激IL-10的分泌,从而抑制树突状细胞中IL-12的表达。MDSC也通过细胞表面的TGFβ和NK受体p30诱导NK细胞失活。此外,MDSC还通过诱导CD4+CD25+叉头转录因子3+调节性T淋巴细胞的产生来抑制免疫反应从而促进了HCC的发生发展[33]。
癌症干细胞(CSC)在肿瘤的发生、发展、复发和转移中也具有重要作用。HIF因子可利用各种机制影响CSC的诱导和发育。这些机制可能是通过诱导CSC标记,CD44、ALDH,腺苷/STAT3/IL-6途径,MAPK/ERK途径,NOTCH和Wnt信号传导或其他机制促进CSC的繁殖[34]。缺氧可显著增强HCC细胞的干细胞相关特性,这种作用可以通过敲低HIF-1α或HIF-2α来消除。此外,HIF-1α特异性干扰RNA处理,在RNA和蛋白质水平显著降低CSC中CD133的表达。重要的是,EMT激活可以诱CSC特征。Notch1通过与HIF-1α的直接相互作用介导EMT诱导的CSC的过程;HIF-1α上调Notch的细胞内表达可激活EMT并诱导HCC细胞在体外获得CSC的特征。
迄今为止,大多数研究报道了HIF-1α和HIF-2α对肿瘤生长的积极作用。然而,另有研究[35]显示,HIF在一些其他癌症中表现出相反的影响,如缺氧可降低野生型(HIF-1α+/+)胚胎干细胞的增殖并增加细胞凋亡。因此,尚需更多研究探析HIF对HCC生长迁移的具体作用。
总结HIF在HCC中的作用机制见图 1。
3. HIF在HCC中的临床意义
3.1 HIF在HCC中的治疗应用
HIF抑制剂有望成为一种有效的HCC治疗方法,抑制HIF-1α活化可能有助于阻止癌症进展,从而使生长的肿瘤细胞缺乏O2和所需的营养供应。当前已发现不同的化合物或药物可通过不同的分子机制阻断HIF的活性,包括减少HIF-1α的蛋白合成,代表药物如mTOR抑制剂、强心苷、拓扑异构酶抑制剂和合成寡核苷酸等;降低HIF-1α mRNA的水平,如前药AFP-464的氨基黄酮成分药物;增加HIF-1α的分解,如HSP90抑制剂、抗氧化剂和硒甲基硒代半胱氨酸药物;减少HIF亚基的异二聚化,如吖啶黄药物;减少DNA与HIF的结合,并降低转录活性,如蒽环类和棘霉素等[36]。研究[37-38]显示,一些中药方剂或中草药提取物也具有抑制或降低HIF-1α的作用,如益气化瘀解毒方、水飞蓟的提取物——水飞蓟素等。
HIF也可能参与了HCC化疗药物耐药性的发展。索拉非尼是一种具有抗增殖、抗血管生成和促凋亡特性的多激酶抑制剂,但由于产生耐药细胞,其疗效不佳,可能是长期索拉非尼治疗后导致补偿性生长途径的激活,或者缺氧诱导的自噬,从而使HIF介导的细胞反应触发对缺氧微环境的适应性机制。因此,HIF抑制剂可以与现有疗法联合使用,以增强常规疗法的敏感性和效果[39]。例如,辛伐他汀可通过抑制HIF-1α/PPAR-γ/PKM2轴,使HCC细胞重新对索拉非尼敏感[40]。此外,有研究[41]显示,HIF-1α可通过促进HCC干细胞特性,促进其对表阿霉素化疗药物的耐药。
然而,开展相关临床研究时必须谨慎。例如,由于HIF抑制剂可能加剧胰岛素抵抗损伤,并抑制肝再生。因此,对于因行肝脏手术的HCC患者,应停止HIF抑制剂治疗[42]。
3.2 HIF在HCC预后评估中的应用
HIF可通过多种途径影响HCC的发生发展,也因此被纳入HCC预后评估的相关研究中。有研究[43]显示,HIF-1α相关基因的过度表达与转移和病死率有关,HIF-1α阳性表达与HCC合并肝硬化患者血管浸润、TNM分期、HBV感染、肿瘤大小、门静脉肿瘤血栓显著相关。如上所述,HIF通过不同方式影响了HCC的发病和进展。因此,HIF相关指标水平的测定有望为HCC预后评估提供价值。
KIAA1199是一种与癌症转移相关的蛋白质,其与HIF-1α在许多人类癌症中上调,机制上与血管浸润、肿瘤TNM分期、HBV感染、肿瘤大小和肝硬化显著相关,提示KIAA1199联合HIF-1α检测对评估HCC患者的存活时间、预后情况有潜在价值[44]。其他如YTHDF1、DAAM2、H3K9me2、H3K9me3、Rpn10等作为HCC预后标志物也被相关研究[45-48]初步证实了临床价值,这些标志物联合HIF-1α检测或许能够作为评估HCC预后的新方法。
4. 讨论与展望
总结现有研究,HIF在HCC中的主要作用是通过促进糖酵解、血管生成、侵袭转移、免疫逃避、癌症干细胞等方式,影响HCC的发生发展以及对治疗耐药,也由此引出了通过HIF靶向治疗HCC和评估HCC预后的新设想。然而,HIF在HCC中的相关研究仍有诸多值得深入探究的问题:(1)既往研究主要着眼于HIF-1,HIF-2次之,而HIF-3的相关研究甚少。已有证据[49]显示,HIF3 AmRNA被差异剪接以产生多种HIF-3α变体,这些变体促进或抑制其他HIF复合物的活性,不同的HIF-3变体具有不同甚至相反的功能。(2)已有研究提供了关于HIF-2α在HCC生长和进展中的稳定性、转录活性和作用的信息,但在HCC中的确切作用尚存争议。有研究[50]认为HIF-2α在肿瘤中可能具有某些积极作用。(3)HIF检测评估预后也存在争议,且尚不完全清楚HIF-1、HIF-2、HIF-3三者在HCC中存在哪些相互作用。相关研究[51]发现,单独敲除HIF-1α或HIF-2α未能显著减小肿瘤体积(HIF-1α敲除后HIF-2α的表达增加,HIF-2α敲除后HIF-1α的表达增加),而同时敲除HIF-1α和HIF-2α之后效果可见肿瘤体积显著缩小。因此,HIF-1、HIF-2、HIF-3之间是否存在一些相互调控作用及其具体机制?如果存在相互调控作用,靶向HIF-1α治疗和HIF-1α评估预后的同时,是否要考虑HIF-2和HIF-3的影响?这些问题有待更多研究探明,以提升相关药物研发的成功率及HIF应用于临床的可靠性。
-
[1] CRAIG AJ, VON FELDEN J, GARCIA-LEZANA T, et al. Tumour evolution in hepatocellular carcinoma[J]. Nat Rev Gastroenterol Hepatol, 2020, 17(3): 139-152. DOI: 10.1038/s41575-019-0229-4. [2] BRAY F, FERLAY J, SOERJOMATARAM I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2018, 68(6): 394-424. DOI: 10.3322/caac.21492. [3] BRUIX J, REIG M, SHERMAN M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma[J]. Gastroenterology, 2016, 150(4): 835-853. DOI: 10.1053/j.gastro.2015.12.041. [4] WU Y, LIU Z, XU X. Molecular subtyping of hepatocellular carcinoma: A step toward precision medicine[J]. Cancer Commun (Lond), 2020, 40(12): 681-693. DOI: 10.1002/cac2.12115. [5] GONG J, CHEHRAZI-RAFFLE A, REDDI S, et al. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: A comprehensive review of registration trials and future considerations[J]. J Immunother Cancer, 2018, 6(1): 8. DOI: 10.1186/s40425-018-0316-z. [6] LLOVET JM, ZUCMAN-ROSSI J, PIKARSKY E, et al. Hepatocellular carcinoma[J]. Nat Rev Dis Primers, 2016, 2: 16018. DOI: 10.1038/nrdp.2016.18. [7] LLOVET JM, BRU' C, BRUIX J. Prognosis of hepatocellular carcinoma: The BCLC staging classification[J]. Semin Liver Dis, 1999, 19(3): 329-338. DOI: 10.1055/s-2007-1007122. [8] BANNASCH P, RIBBACK S, SU Q, et al. Clear cell hepatocellular carcinoma: Origin, metabolic traits and fate of glycogenotic clear and ground glass cells[J]. Hepatobiliary Pancreat Dis Int, 2017, 16(6): 570-594. DOI: 10.1016/S1499-3872(17)60071-7. [9] FAIVRE S, RIMASSA L, FINN RS. Molecular therapies for HCC: Looking outside the box[J]. J Hepatol, 2020, 72(2): 342-352. DOI: 10.1016/j.jhep.2019.09.010. [10] ERSTAD DJ, FUCHS BC, TANABE KK. Molecular signatures in hepatocellular carcinoma: A step toward rationally designed cancer therapy[J]. Cancer, 2018, 124(15): 3084-3104. DOI: 10.1002/cncr.31257. [11] CHIANG DY, VILLANUEVA A, HOSHIDA Y, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma[J]. Cancer Res, 2008, 68(16): 6779-6788. DOI: 10.1158/0008-5472.CAN-08-0742. [12] SCHULZE K, IMBEAUD S, LETOUZÉ E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets[J]. Nat Genet, 2015, 47(5): 505-511. DOI: 10.1038/ng.3252. [13] FUJIMOTO A, FURUTA M, TOTOKI Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer[J]. Nat Genet, 2016, 48(5): 500-509. DOI: 10.1038/ng.3547. [14] LEE JS, CHU JS, CHU IS, HEO J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling[J]. Hepatology, 2004, 40(3): 667-676. DOI: 10.1002/hep.20375. [15] JIANG X, KIM HE, SHU H, et al. Distinctive roles of PHAP proteins and prothymosin-alpha in a death regulatory pathway[J]. Science, 2003, 299(5604): 223-226. DOI: 10.1126/science.1076807. [16] SEMENZA GL. Targeting HIF-1 for cancer therapy[J]. Nat Rev Cancer, 2003, 3(10): 721-732. DOI: 10.1038/nrc1187. [17] LEE JS, HEO J, LIBBRECHT L, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells[J]. Nat Med, 2006, 12(4): 410-416. DOI: 10.1038/nm1377. [18] BOYAULT S, RICKMAN DS, de REYNIÈS A, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets[J]. Hepatology, 2007, 45(1): 42-52. DOI: 10.1002/hep.21467. [19] HOSHIDA Y, NIJMAN SM, KOBAYASHI M, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma[J]. Cancer Res, 2009, 69(18): 7385-7392. DOI: 10.1158/0008-5472.CAN-09-1089. [20] ZAVADIL J, BÖTTINGER EP. TGF-beta and epithelial-to-mesenchymal transitions[J]. Oncogene, 2005, 24(37): 5764-5774. DOI: 10.1038/sj.onc.1208927. [21] GIANNELLI G, BERGAMINI C, FRANSVEA E, et al. Laminin-5 with transforming growth factor-beta1 induces epithelial to mesenchymal transition in hepatocellular carcinoma[J]. Gastroenterology, 2005, 129(5): 1375-1383. DOI: 10.1053/j.gastro.2005.09.055. [22] HOPPLER S, KAVANAGH CL. Wnt signalling: Variety at the core[J]. J Cell Sci, 2007, 120(Pt 3): 385-393. DOI: 10.1242/jcs.03363. [23] MUTO Y, MORIWAKI H, SHIRATORI Y. Prevention of second primary tumors by an acyclic retinoid, polyprenoic acid, in patients with hepatocellular carcinoma[J]. Digestion, 1998, 59(Suppl 2): 89-91. DOI: 10.1159/000051435. [24] ZHANG Y, WANG G, MA W, et al. CdS p-n heterojunction co-boosting with Co(3)O(4) and Ni-MOF-74 for photocatalytic hydrogen evolution[J]. Dalton Trans, 2018, 47(32): 11176-11189. DOI: 10.1039/c8dt02294a. [25] HANAHAN D, WEINBERG RA. Hallmarks of cancer: The next generation[J]. Cell, 2011, 144(5): 646-674. DOI: 10.1016/j.cell.2011.02.013. [26] DÉSERT R, ROHART F, CANAL F, et al. Human hepatocellular carcinomas with a periportal phenotype have the lowest potential for early recurrence after curative resection[J]. Hepatology, 2017, 66(5): 1502-1518. DOI: 10.1002/hep.29254. [27] YANG C, HUANG X, LIU Z, et al. Metabolism-associated molecular classification of hepatocellular carcinoma[J]. Mol Oncol, 2020, 14(4): 896-913. DOI: 10.1002/1878-0261.12639. [28] SHANKARAIAH RC, CALLEGARI E, GUERRIERO P, et al. Metformin prevents liver tumourigenesis by attenuating fibrosis in a transgenic mouse model of hepatocellular carcinoma[J]. Oncogene, 2019, 38(45): 7035-7045. DOI: 10.1038/s41388-019-0942-z. [29] DUFFY AG, ULAHANNAN SV, MAKOROVA-RUSHER O, et al. Tremelimumab in combination with ablation in patients with advanced hepatocellular carcinoma[J]. J Hepatol, 2017, 66(3): 545-551. DOI: 10.1016/j.jhep.2016.10.029. [30] EL-KHOUEIRY AB, SANGRO B, YAU T, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): An open-label, non-comparative, phase 1/2 dose escalation and expansion trial[J]. Lancet, 2017, 389(10088): 2492-2502. DOI: 10.1016/S0140-6736(17)31046-2. [31] KILLOCK D. Immunotherapy: Nivolumab keeps HCC in check and opens avenues for checkmate[J]. Nat Rev Clin Oncol, 2017, 14(7): 392. DOI: 10.1038/nrclinonc.2017.70. [32] JIANG Y, SUN A, ZHAO Y, et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma[J]. Nature, 2019, 567(7747): 257-261. DOI: 10.1038/s41586-019-0987-8. [33] GAO Q, ZHU H, DONG L, et al. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma[J]. Cell, 2019, 179(2): 561-577. e22. DOI: 10.1016/j.cell.2019.08.052. [34] SIA D, JIAO Y, MARTINEZ-QUETGLAS I, et al. Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features[J]. Gastroenterology, 2017, 153(3): 812-826. DOI: 10.1053/j.gastro.2017.06.007. [35] KUREBAYASHI Y, OJIMA H, TSUJIKAWA H, et al. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification[J]. Hepatology, 2018, 68(3): 1025-1041. DOI: 10.1002/hep.29904. [36] KIM HD, SONG GW, PARK S, et al. Association between expression level of PD1 by tumor-infiltrating CD8+T cells and features of hepatocellular carcinoma[J]. Gastroenterology, 2018, 155(6): 1936-1950. e17. DOI: 10.1053/j.gastro.2018.08.030. [37] ZHANG Q, LOU Y, YANG J, et al. Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas[J]. Gut, 2019, 68(11): 2019-2031. DOI: 10.1136/gutjnl-2019-318912. [38] LIM HY, HEO J, CHOI HJ, et al. A phase Ⅱ study of the efficacy and safety of the combination therapy of the MEK inhibitor refametinib (BAY 86-9766) plus sorafenib for Asian patients with unresectable hepatocellular carcinoma[J]. Clin Cancer Res, 2014, 20(23): 5976-5985. DOI: 10.1158/1078-0432.CCR-13-3445. [39] HO DWH, CHAN LK, CHIU YT, et al. TSC1/2 mutations define a molecular subset of HCC with aggressive behaviour and treatment implication[J]. Gut, 2017, 66(8): 1496-1506. DOI: 10.1136/gutjnl-2016-312734. [40] SUN W, LI SC, XU L, et al. High FLT3 levels may predict sorafenib benefit in hepatocellular carcinoma[J]. Clin Cancer Res, 2020, 26(16): 4302-4312. DOI: 10.1158/1078-0432.CCR-19-1858. 期刊类型引用(4)
1. 范智博,魏绵兴,李胜鸿,徐晓梅. 低氧条件下HIF-1α对骨生理中成骨和破骨的影响. 西南医科大学学报. 2024(01): 80-86 .  百度学术
百度学术2. 李香香,王振,杨星,李素领. 中药抑制肝细胞癌血管生成的作用机制. 临床肝胆病杂志. 2024(07): 1477-1485 .  本站查看
本站查看3. 黄丽恒,林洪升,邓增富,杨利,李明芬. 缺氧诱导因子1α在肝细胞癌靶点治疗中的研究进展. 实用临床医药杂志. 2024(17): 127-130 .  百度学术
百度学术4. 陈纪宏,许向波,何福亮,田苗苗,胡思隽,祁兴顺. 缺氧环境下的肝星状细胞中缺氧诱导因子1α与缺氧诱导因子2α对CD248表达水平影响. 临床军医杂志. 2023(09): 884-891 .  百度学术
百度学术其他类型引用(0)
-




 PDF下载 ( 2840 KB)
PDF下载 ( 2840 KB)


 下载:
下载:
 下载:
下载:
 百度学术
百度学术