Screening of differentially expressed circular RNAs in intrahepatic cholangiocarcinoma based on microarray technique and potential mechanism of circRNA_000585
-
摘要:
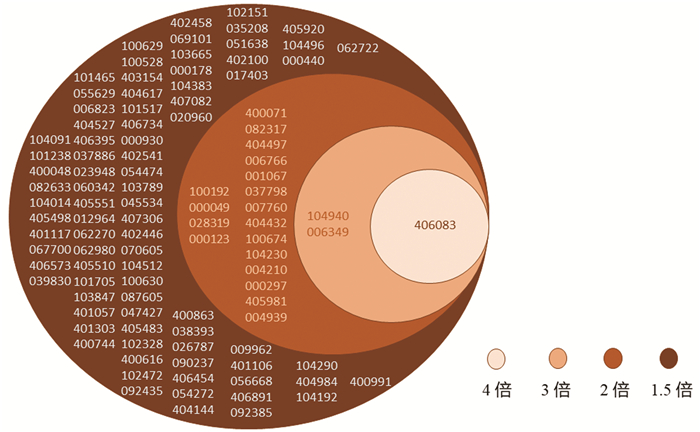
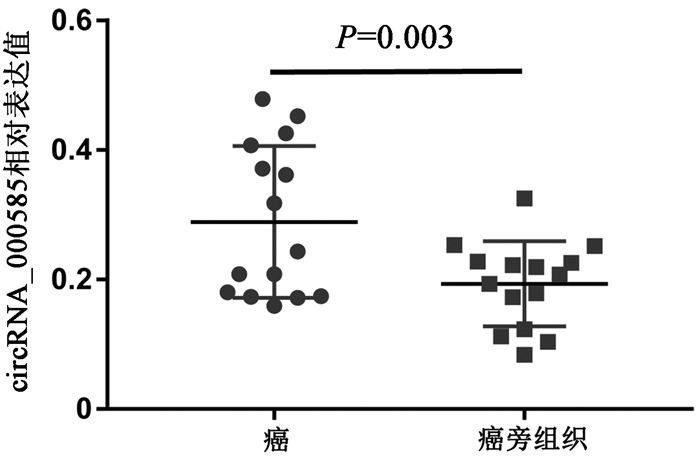
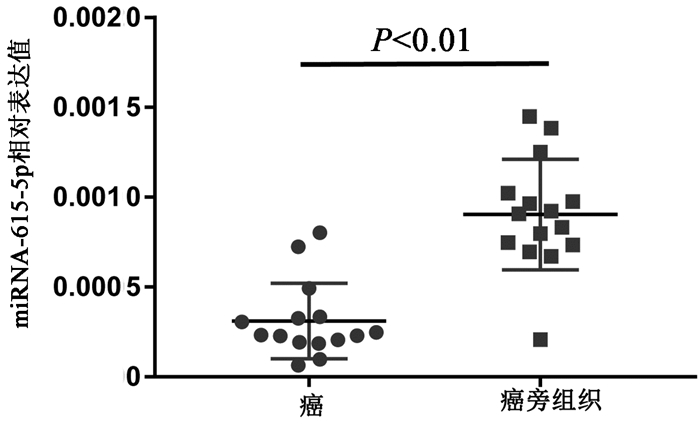
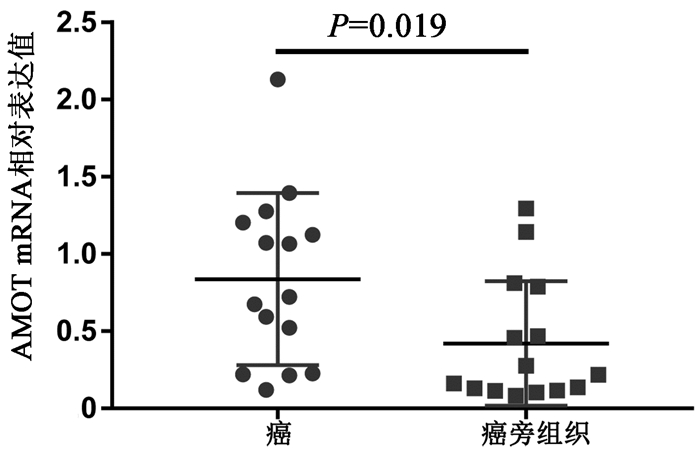
目的 通过对肝内胆管细胞癌(iCCA)和配对的癌旁组织的circRNA芯片比较分析,发现并验证iCCA中异常表达的circRNA,进而揭示其潜在的机制。 方法 收集2019年7月—12月收治的3例iCCA患者肿瘤组织标本,采用微阵列芯片杂交的方法检测iCCA和癌旁组织中circRNA的差异表达,并收集同一时期15例iCCA患者样本,采用实时定量PCR(RT-PCR)验证差异表达的circRNA。通过生物信息学分析寻找差异表达的circRNA的下游分子,并进一步采用RT-PCR对潜在分子进行验证。计量资料组间比较采用配对样本t检验;计数资料组间比较采用χ2检验或Fisher精确检验。 结果 采用1.5倍作为差异表达界值,结果发现iCCA相比癌旁组织中117个circRNAs出现上调,104个circRNAs出现下调。采用3倍作为界值,发现iCCA中10个circRNAs出现上调(circRNA_002172、circRNA_002144、circRNA_001588、circRNA_000166、circRNA_000585、circRNA_000167、circRNA_402608、circRNA_006853、circRNA_001589、circRNA_008882),3个circRNAs出现下调(circRNA_406083、circRNA_104940、circRNA_006349)。收集15例病理证实的iCCA及癌旁组织,采用RT-PCR进行验证差异表达的circRNA,结果发现circRNA_000585在iCCA中表达显著升高(t=3.607,P=0.003)。进一步通过生物信息学分析发现circRNA_000585/miR-615-5p/AMOT/YAP可能是iCCA发生的潜在通路,RT-PCR验证发现该通路中miR-615-5p在iCCA中显著下调(t=5.724,P<0.001),而AMOT和YAP分子在iCCA中显著上调(t值分别为2.664、2.986,P值分别为0.019、0.009 8)。 结论 多种circRNAs在iCCA中出现异常表达,其中circRNA_000585在iCCA中显著上调,其可能通过circRNA_000585/miR-615-5p/AMOT/YAP信号通路在iCCA的发生中发挥重要作用。 Abstract:Objective To screen out and validate the abnormally expressed circular RNAs (circRNAs) in intrahepatic cholangiocarcinoma (iCCA) by comparing the circRNA microarray results of iCCA tissue and adjacent tissue, and to investigate their potential mechanism in iCCA. Methods Tumor tissue specimens were collected from three patients with iCCA who were admitted from July to December, 2019, and microarray hybridization was used to measure the differential expression of circRNAs between iCCA tissue and adjacent tissue. A total of 15 patients with iCCA who were treated during the same period of time were enrolled, and quantitative real-time PCR was used for validation of differentially expressed circRNAs. A bioinformatics analysis was performed to identify the downstream molecules of differentially expressed circRNAs, and quantitative real-time PCR was used for validation of potential molecules. The paired samples t-test was used for comparison of continuous data between groups, and the chi-square test or the Fisher's exact test was used for comparison of categorical data between groups. Results With 1.5-fold as the cut-off value for differential expression, there were 171 upregulated circRNAs and 104 downregulated circRNAs in iCCA tissue compared with the adjacent tissue. With 3-fold as the cut-off value for differential expression, there were 10 upregulated circRNAs (circRNA_002172, circRNA_002144, circRNA_001588, circRNA_000166, circRNA_000585, circRNA_000167, circRNA_402608, circRNA_006853, circRNA_001589, and circRNA_008882) and 3 downregulated circRNAs (circRNA_406083, circRNA_104940, and circRNA_006349) compared with the adjacent tissue. Pathologically confirmed iCCA tissue samples and adjacent tissue samples were collected from 15 patients, and quantitative real-time PCR was used for the validation of differentially expressed circRNAs; the results showed that circRNA_000585 was significantly upregulated in iCCA tissue (t=3.607, P=0.003). Further bioinformatics analysis showed that circRNA_000585/miR-615-5p/AMOT/YAP might be the potential pathway involved in the pathogenesis of iCCA, and quantitative real-time PCR showed that for this pathway, miR-615-5p was significantly downregulated in iCCA (t=5.724, P < 0.001) and AMOT and YAP were significantly upregulated in iCCA (t=2.664 and 2.986, P=0.019 and 0.009 8). Conclusion Abnormal expression of various circRNAs is observed in iCCA, among which circRNA_000585 is significantly upregulated in iCCA and may play an important role in the pathogenesis of iCCA via the circRNA_000585/miR-615-5p/AMOT/YAP pathway. -
Key words:
- Cholangiocarcinoma /
- Gene Expression /
- Computational Biology
-
表 1 circRNA_000585和内参PCR信息
基因名称 引物(5′-3′) 退火温度(℃) 扩增产物长度(bp) 18S rRNA F: CAGCCACCCGAGATTGAGCA R: TAGTAGCGACGGGCGGTGTG 60 252 hsa_circRNA_000585 F: GAGGTCAGACTGGGCAGGAGAT R: ACAGGACGCACTCAGTTCGCT 60 117 表 2 miR-615-5p和内参PCR信息
基因名称 引物(5′-3′) 退火温度(℃) 扩增产物长度(bp) U6 F: GCTTCGGCAGCACATATACTAAAAT R: CGCTTCACGAATTTGCGTGTCAT 60 89 hsa-miR-615-5p GSP: AAGGGGGTCCCCGGT 60 62 R: GTGCGTGTCGTGGAGTCG 表 3 AMOT/YAP和内参PCR信息
基因名称 引物(5′-3′) 退火温度(℃) 扩增产物长度(bp) AMOT F: TTGGAGGAGAATGTGATGAGAC R: TGGTGTTAGGAGAGTGACTGATG 60 97 YAP F: GCCAGCAGGTTGGGAGAT R: TGTGATTTAAGAAGTATCTCTGACC 60 59 18S rRNA F: CAGCCACCCGAGATTGAGCA R: TAGTAGCGACGGGCGGTGTG 60 252 -
[1] RIZVI S, KHAN SA, HALLEMEIER CL, et al. Cholangiocarcinoma-evolving concepts and therapeutic strategies[J]. Nat Rev Clin Oncol, 2018, 15(2): 95-111. DOI: 10.1038/nrclinonc.2017.157. [2] VALLE J, WASAN H, PALMER DH, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer[J]. N Engl J Med, 2010, 362(14): 1273-1281. DOI: 10.1056/NEJMoa0908721. [3] LABIB PL, GOODCHILD G, PEREIRA SP. Molecular pathogenesis of cholangiocarcinoma[J]. BMC Cancer, 2019, 19(1): 185. DOI: 10.1186/s12885-019-5391-0. [4] LI J, SUN D, PU W, et al. Circular RNAs in cancer: Biogenesis, function, and clinical significance[J]. Trends Cancer, 2020, 6(4): 319-336. DOI: 10.1016/j.trecan.2020.01.012. [5] JIANG XM, LI ZL, LI JL, et al. A novel prognostic biomarker for cholangiocarcinoma: circRNA Cdr1as[J]. Eur Rev Med Pharmacol Sci, 2018, 22(2): 365-371. DOI: 10.26355/eurrev_201801_14182. [6] LU Q, FANG T. Circular RNA SMARCA5 correlates with favorable clinical tumor features and prognosis, and increases chemotherapy sensitivity in intrahepatic cholangiocarcinoma[J]. J Clin Lab Anal, 2020, 34(4): e23138. DOI: 10.1002/jcla.23138. [7] LI D, TANG Z, GAO Z, et al. Circular RNA CDR1as exerts oncogenic properties partially through regulating MicroRNA 641 in cholangiocarcinoma[J]. Mol Cell Biol, 2020, 40(15). DOI: 10.1128/MCB.00042-20. [8] LOUIS C, DESOTEUX M, COULOUARN C. Exosomal circRNAs: New players in the field of cholangiocarcinoma[J]. Clin Sci (Lond), 2019, 133(21): 2239-2244. DOI: 10.1042/CS20190940. [9] MOIRANGTHEM A, WANG X, YAN IK, et al. Network analyses- based identification of circular ribonucleic acid-related pathways in intrahepatic cholangiocarcinoma[J]. Tumour Biol, 2018, 40(9): 1010428318795761. DOI: 10.1177/1010428318795761. [10] WANG S, HU Y, LV X, et al. Circ-0000284 arouses malignant phenotype of cholangiocarcinoma cells and regulates the biological functions of peripheral cells through cellular communication[J]. Clin Sci (Lond), 2019, 133(18): 1935-1953. DOI: 10.1042/CS20190589. [11] XU Y, YAO Y, ZHONG X, et al. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells[J]. Biochem Biophys Res Commun, 2018, 496(2): 455-461. DOI: 10.1016/j.bbrc.2018.01.077. [12] XU Y, YAO Y, LIU Y, et al. Elevation of circular RNA circ_0005230 facilitates cell growth and metastasis via sponging miR-1238 and miR-1299 in cholangiocarcinoma[J]. Aging (Albany NY), 2019, 11(7): 1907-1917. DOI: 10.18632/aging.101872. [13] GUO S, XU X, OUYANG Y, et al. Microarray expression profile analysis of circular RNAs in pancreatic cancer[J]. Mol Med Rep, 2018, 17(6): 7661-7671. DOI: 10.3892/mmr.2018.8827. [14] MUMTAZ PT, TABAN Q, DAR MA, et al. Deep insights in circular RNAs: From biogenesis to therapeutics[J]. Biol Proced Online, 2020, 22: 10. DOI: 10.1186/s12575-020-00122-8. [15] SONG H, LIU Q, LIAO Q. Circular RNA and tumor microenvironment[J]. Cancer Cell Int, 2020, 20: 211. DOI: 10.1186/s12935-020-01301-z. [16] LIANG ZZ, GUO C, ZOU MM, et al. circRNA-miRNA-mRNA regulatory network in human lung cancer: An update[J]. Cancer Cell Int, 2020, 20: 173. DOI: 10.1186/s12935-020-01245-4. [17] GODÍNEZ-RUBÍ M, ORTUÑO-SAHAGÚN D. miR-615 fine-tunes growth and development and has a role in cancer and in neural repair[J]. Cells, 2020, 9(7): 1566. DOI: 10.3390/cells9071566. [18] HUANG T, ZHOU Y, ZHANG J, et al. The physiological role of Motin family and its dysregulation in tumorigenesis[J]. J Transl Med, 2018, 16(1): 98. DOI: 10.1186/s12967-018-1466-y. [19] HONG W. Angiomotin'g YAP into the nucleus for cell proliferation and cancer development[J]. Sci Signal, 2013, 6(291): pe27. DOI: 10.1126/scisignal.2004573. [20] LV M, SHEN Y, YANG J, et al. Angiomotin family members: Oncogenes or tumor suppressors?[J]. Int J Biol Sci, 2017, 13(6): 772-781. DOI: 10.7150/ijbs.19603. [21] POCATERRA A, ROMANI P, DUPONT S. YAP/TAZ functions and their regulation at a glance[J]. J Cell Sci, 2020, 133(2): jcs230425. DOI: 10.1242/jcs.230425. [22] MA S, MENG Z, CHEN R, et al. The hippo pathway: Biology and pathophysiology[J]. Annu Rev Biochem, 2019, 88: 577-604. DOI: 10.1146/annurev-biochem-013118-111829. [23] SHAO DD, XUE W, KRALL EB, et al. KRAS and YAP1 converge to regulate EMT and tumor survival[J]. Cell, 2014, 158(1): 171-184. DOI: 10.1016/j.cell.2014.06.004. [24] YAMAGUCHI H, TAOUK GM. A potential role of YAP/TAZ in the interplay between metastasis and metabolic alterations[J]. Front Oncol, 2020, 10: 928. DOI: 10.3389/fonc.2020.00928. [25] MARTI P, STEIN C, BLUMER T, et al. YAP promotes proliferation, chemoresistance, and angiogenesis in human cholangiocarcinoma through TEAD transcription factors[J]. Hepatology, 2015, 62(5): 1497-1510. DOI: 10.1002/hep.27992. [26] PARK J, KIM JS, NAHM JH, et al. WWC1 and NF2 prevent the development of intrahepatic cholangiocarcinoma by regulating YAP/TAZ activity through LATS in mice[J]. Mol Cells, 2020, 43(5): 491-499. DOI: 10.14348/molcells.2020.0093. 期刊类型引用(3)
1. 梁海锋,梁国志,袁观斗,郭振亚,何松青. 小肝综合征的发病机制及防治研究进展. 中华实验外科杂志. 2024(03): 639-643 .  百度学术
百度学术2. 蔡景治,郑扬,仲伟明,刘波. 基于Bcl-2/CytC探究槲皮素对肝硬化大鼠微循环及肝脏病变的影响. 现代生物医学进展. 2023(05): 818-823 .  百度学术
百度学术3. 赵媛,赵明,范玉梅,邵建营. 失代偿期乙型肝炎肝硬化患者不同病毒学应答状态对生存率及肝癌患病率的影响. 中国实用医刊. 2020(23): 50-52 .  百度学术
百度学术其他类型引用(0)
-




 PDF下载 ( 3552 KB)
PDF下载 ( 3552 KB)


 下载:
下载:



 百度学术
百度学术

