| [1] |
SWANSON KS, de VOS WM, MARTENS EC, et al. Effect of fructans, prebiotics and fibres on the human gut microbiome assessed by 16S rRNA-based approaches: A review[J]. Benef Microbes, 2020, 11( 2): 101- 129. DOI: 10.3920/BM2019.0082. |
| [2] |
LI ZQ, CHU JT, SU FX, et al. Characteristics of bile microbiota in cholelithiasis, perihilar cholangiocarcinoma, distal cholangiocarcinoma, and pancreatic cancer[J]. Am J Transl Res, 2022, 14( 5): 2962- 2971.
|
| [3] |
SINAGRA E, UTZERI E, MORREALE GC, et al. Microbiota-gut-brain axis and its affect inflammatory bowel disease: Pathophysiological concepts and insights for clinicians[J]. World J Clin Cases, 2020, 8( 6): 1013- 1025. DOI: 10.12998/wjcc.v8.i6.1013. |
| [4] |
NASCIMENTO FSD, SUZUKI MO, TABA JV, et al. Analysis of biliary MICRObiota in hepatoBILIOpancreatic diseases compared to healthy people[MICROBILIO]: Study protocol[J]. PLoS One, 2020, 15( 11): e0242553. DOI: 10.1371/journal.pone.0242553. |
| [5] |
BOYER JL. Bile formation and secretion[J]. Compr Physiol, 2013, 3( 3): 1035- 1078. DOI: 10.1002/cphy.c120027. |
| [6] |
CSENDES A, BURDILES P, MALUENDA F, et al. Simultaneous bacteriologic assessment of bile from gallbladder and common bile duct in control subjects and patients with gallstones and common duct stones[J]. Arch Surg, 1996, 131( 4): 389- 394. DOI: 10.1001/archsurg.1996.01430160047008. |
| [7] |
JIMÉNEZ E, SÁNCHEZ B, FARINA A, et al. Characterization of the bile and gall bladder microbiota of healthy pigs[J]. Microbiologyopen, 2014, 3( 6): 937- 949. DOI: 10.1002/mbo3.218. |
| [8] |
MOLINERO N, RUIZ L, MILANI C, et al. The human gallbladder microbiome is related to the physiological state and the biliary metabolic profile[J]. Microbiome, 2019, 7( 1): 100. DOI: 10.1186/s40168-019-0712-8. |
| [9] |
BERG G, RYBAKOVA D, FISCHER D, et al. Correction to: Microbiome definition re-visited: Old concepts and new challenges[J]. Microbiome, 2020, 8( 1): 119. DOI: 10.1186/s40168-020-00905-x. |
| [10] |
WU T, ZHANG ZG, LIU B, et al. Gut microbiota dysbiosis and bacterial community assembly associated with cholesterol gallstones in large-scale study[J]. BMC Genomics, 2013, 14: 669. DOI: 10.1186/1471-2164-14-669. |
| [11] |
YE FQ, SHEN HZ, LI Z, et al. Influence of the biliary system on biliary bacteria revealed by bacterial communities of the human biliary and upper digestive tracts[J]. PLoS One, 2016, 11( 3): e0150519. DOI: 10.1371/journal.pone.0150519. |
| [12] |
SCHNEIDER J, DE WAHA P, HAPFELMEIER A, et al. Risk factors for increased antimicrobial resistance: A retrospective analysis of 309 acute cholangitis episodes[J]. J Antimicrob Chemother, 2014, 69( 2): 519- 525. DOI: 10.1093/jac/dkt373. |
| [13] |
SUNG JY, COSTERTON JW, SHAFFER EA. Defense system in the biliary tract against bacterial infection[J]. Dig Dis Sci, 1992, 37( 5): 689- 696. DOI: 10.1007/BF01296423. |
| [14] |
LEE J, PARK JS, BAE J, et al. Bile microbiome in patients with recurrent common bile duct stones and correlation with the duodenal microbiome[J]. Life(Basel), 2022, 12( 10): 1540. DOI: 10.3390/life12101540. |
| [15] |
KIM MH, MYUNG SJ, SEO DW, et al. Association of periampullary diverticula with primary choledocholithiasis but not with secondary choledocholithiasis[J]. Endoscopy, 1998, 30( 7): 601- 604. DOI: 10.1055/s-2007-1001363. |
| [16] |
LAI KH, PENG NJ, LO GH, et al. Prediction of recurrent choledocholithiasis by quantitative cholescintigraphy in patients after endoscopic sphincterotomy[J]. Gut, 1997, 41( 3): 399- 403. DOI: 10.1136/gut.41.3.399. |
| [17] |
CHEN M, WANG L, WANG Y, et al. Risk factor analysis of post-ERCP cholangitis: A single-center experience[J]. Hepatobiliary Pancreat Dis Int, 2018, 17( 1): 55- 58. DOI: 10.1016/j.hbpd.2018.01.002. |
| [18] |
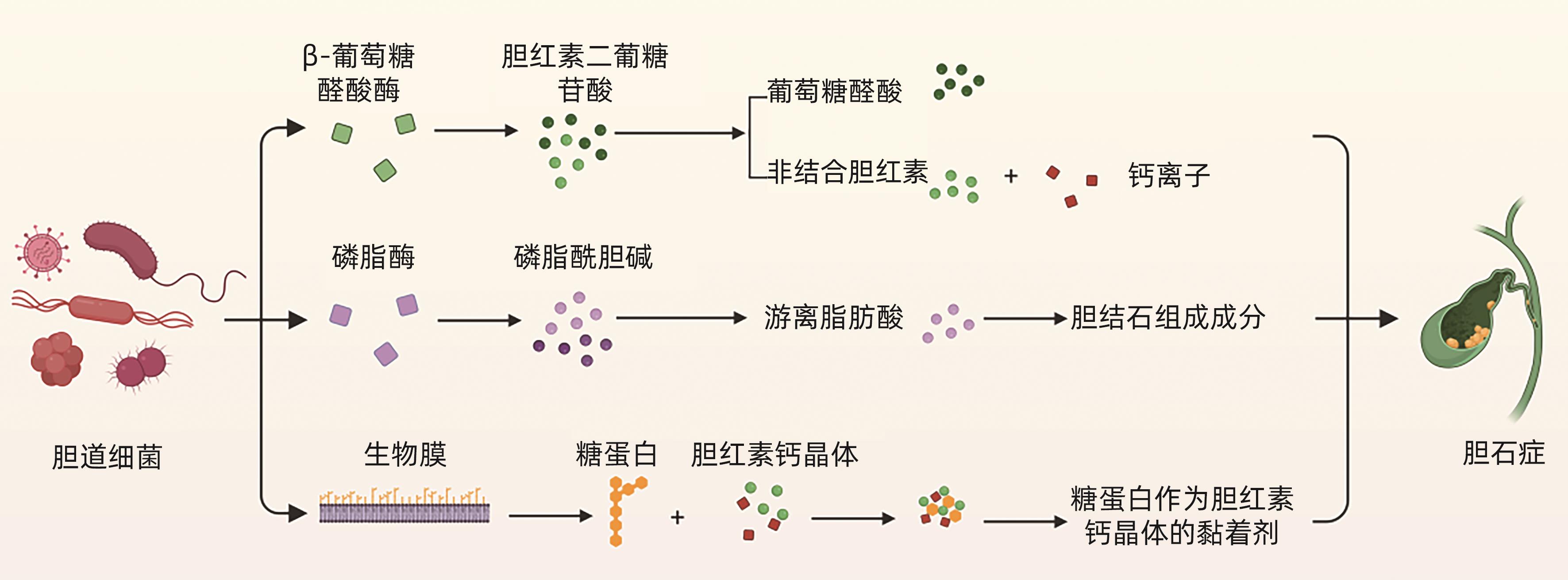
MAKI T. Pathogenesis of calcium bilirubinate gallstone: Role of E. coli, beta-glucuronidase and coagulation by inorganic ions, polyelectrolytes and agitation[J]. Ann Surg, 1966, 164( 1): 90- 100. DOI: 10.1097/00000658-196607000-00010. |
| [19] |
TAJEDDIN E, SHERAFAT SJ, MAJIDI MRS, et al. Association of diverse bacterial communities in human bile samples with biliary tract disorders: A survey using culture and polymerase chain reaction-denaturing gradient gel electrophoresis methods[J]. Eur J Clin Microbiol Infect Dis, 2016, 35( 8): 1331- 1339. DOI: 10.1007/s10096-016-2669-x. |
| [20] |
HUANG D, WAN XJ. Research progress in the formation mechanism of bile duct stones[J]. Adv Clin Med, 2023, 9( 10): 16950- 16959.
黄丹, 宛新建. 胆管结石形成机制的研究进展[J]. 临床医学进展, 2023, 9( 10): 16950- 16959.
|
| [21] |
HU FL, CHEN HT, GUO FF, et al. Biliary microbiota and mucin 4 impact the calcification of cholesterol gallstones[J]. Hepatobiliary Pancreat Dis Int, 2021, 20( 1): 61- 66. DOI: 10.1016/j.hbpd.2020.12.002. |
| [22] |
NOMURA T, SHIRAI Y, HATAKEYAMA K. Enterococcal bactibilia in patients with malignant biliary obstruction[J]. Dig Dis Sci, 2000, 45( 11): 2183- 2186. DOI: 10.1023/a:1026640603312. |
| [23] |
KIM B, PARK JS, BAE J, et al. Bile microbiota in patients with pigment common bile duct stones[J]. J Korean Med Sci, 2021, 36( 15): e94. DOI: 10.3346/jkms.2021.36.e94. |
| [24] |
CHOE JW, LEE JM, HYUN JJ, et al. Analysis on microbial profiles& components of bile in patients with recurrent CBD stones after endoscopic CBD stone removal: A preliminary study[J]. J Clin Med, 2021, 10( 15): 3303. DOI: 10.3390/jcm10153303. |
| [25] |
GUZIOR DV, QUINN RA. Review: Microbial transformations of human bile acids[J]. Microbiome, 2021, 9( 1): 140. DOI: 10.1186/s40168-021-01101-1. |
| [26] |
BELZER C, KUSTERS JG, KUIPERS EJ, et al. Urease induced calcium precipitation by Helicobacter species may initiate gallstone formation[J]. Gut, 2006, 55( 11): 1678- 1679. DOI: 10.1136/gut.2006.098319. |
| [27] |
SIPOS P, KRISZTINA H, BLÁZOVICS A, et al. Cholecystitis, gallstones and free radical reactions in human gallbladder[J]. Med Sci Monit, 2001, 7( 1): 84- 88.
|
| [28] |
|
| [29] |
HAN JY, WU SD, FAN Y, et al. Biliary microbiota in choledocholithiasis and correlation with duodenal microbiota[J]. Front Cell Infect Microbiol, 2021, 11: 625589. DOI: 10.3389/fcimb.2021.625589. |
| [30] |
LIU J, LIU B, LI HF, et al. Etiological analysis of biliary tract infection[J]. Chin J Nosocomiology, 2018, 28( 20): 3111- 3114. DOI: 10.11816/cn.ni.2018-182836. |
| [31] |
HU FL, SHANG D, ZHANG HX, et al. A review of Tokyo Guidelines 2018 for the management of acute biliary infections[J]. Chin J Pract Surg, 2018, 38( 7): 763- 766. DOI: 10.19538/j.cjps.issn1005-2208.2018.07.15. |
| [32] |
XIONG GZ, ZHANG JF, DAI XP, et al. Bacterial spectrum and antimicrobial resistance of pathogens from bile culture in patients with biliary tract infection[J]. Chin J Nosocomiology, 2012, 22( 1): 198- 199.
熊国祚, 张俊方, 戴先鹏, 等. 胆道感染患者胆汁培养的菌谱调查及耐药性分析[J]. 中华医院感染学杂志, 2012, 22( 1): 198- 199.
|
| [33] |
DALE AP, WOODFORD N. Extra-intestinal pathogenic Escherichia coli(ExPEC): Disease, carriage and clones[J]. J Infect, 2015, 71( 6): 615- 626. DOI: 10.1016/j.jinf.2015.09.009. |
| [34] |
WU ZY, WU XS, YAO WY, et al. Pathogens' distribution and changes of antimicrobial resistance in the bile of acute biliary tract infection patients[J]. Chin J Surg, 2021, 59( 1): 24- 31. DOI: 10.3760/cma.j.cn112139-20200717-00559. |
| [35] |
China Antimicrobial Resistance Surveillance System. Antimicrobial resistance of bacteria from bile: surveillance report from China Antimicrobial Resistance Surveillance System in 2014-2019[J]. Chin J Infect Contr, 2021, 20( 1): 76- 84. DOI: 10.12138/j.issn.1671-9638.20216177. |
| [36] |
MUSSA M, PÉREZ-CRESPO PMM, LOPEZ-CORTES LE, et al. Risk factors and predictive score for bacteremic biliary tract infections due to Enterococcus faecalis and Enterococcus faecium: A multicenter cohort study from the PROBAC project[J]. Microbiol Spectr, 2022, 10( 4): e0005122. DOI: 10.1128/spectrum.00051-22. |
| [37] |
ZHAO CW, LIU SS, BAI X, et al. A retrospective study on bile culture and antibiotic susceptibility patterns of patients with biliary tract infections[J]. Evid Based Complement Alternat Med, 2022, 2022: 9255444. DOI: 10.1155/2022/9255444. |
| [38] |
SARCOGNATO S, SACCHI D, GRILLO F, et al. Autoimmune biliary diseases: Primary biliary cholangitis and primary sclerosing cholangitis[J]. Pathologica, 2021, 113( 3): 170- 184. DOI: 10.32074/1591-951X-245. |
| [39] |
KUMMEN M, THINGHOLM LB, RÜHLEMANN MC, et al. Altered gut microbial metabolism of essential nutrients in primary sclerosing cholangitis[J]. Gastroenterology, 2021, 160( 5): 1784- 1798. DOI: 10.1053/j.gastro.2020.12.058. |
| [40] |
OLSSON R, BJÖRNSSON E, BÄCKMAN L, et al. Bile duct bacterial isolates in primary sclerosing cholangitis: A study of explanted livers[J]. J Hepatol, 1998, 28( 3): 426- 432. DOI: 10.1016/s0168-8278(98)80316-4. |
| [41] |
LIWINSKI T, ZENOUZI R, JOHN C, et al. Alterations of the bile microbiome in primary sclerosing cholangitis[J]. Gut, 2020, 69( 4): 665- 672. DOI: 10.1136/gutjnl-2019-318416. |
| [42] |
HIRAMATSU K, HARADA K, TSUNEYAMA K, et al. Amplification and sequence analysis of partial bacterial 16S ribosomal RNA gene in gallbladder bile from patients with primary biliary cirrhosis[J]. J Hepatol, 2000, 33( 1): 9- 18. DOI: 10.1016/s0168-8278(00)80153-1. |
| [43] |
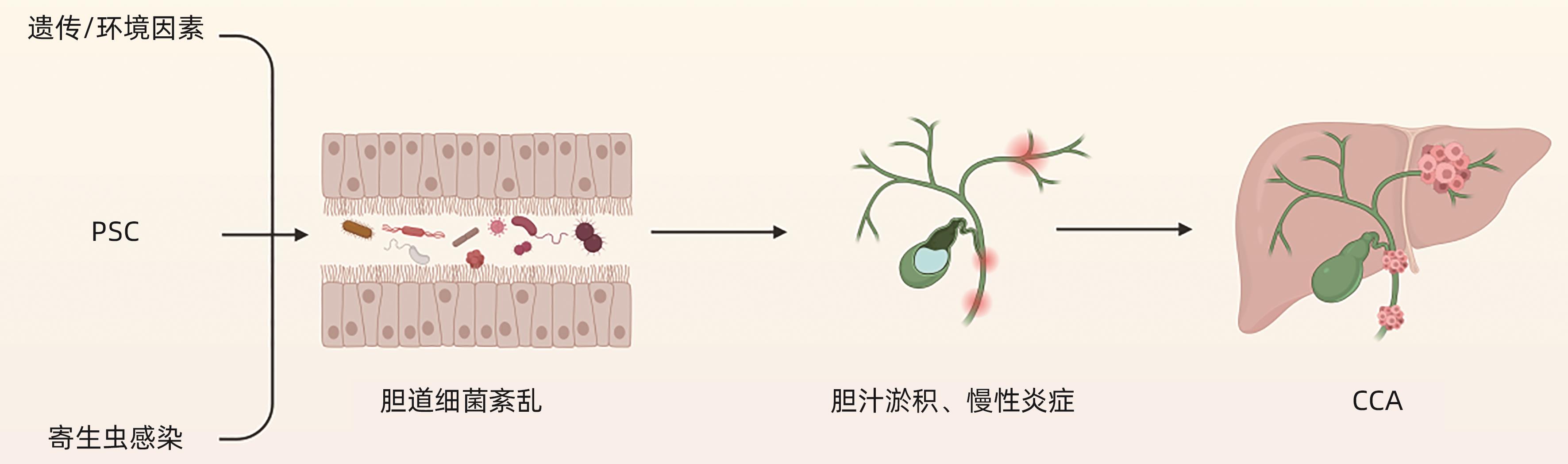
CARDINALE V. Classifications and misclassification in cholangiocarcinoma[J]. Liver Int, 2019, 39( 2): 260- 262. DOI: 10.1111/liv.13998. |
| [44] |
|
| [45] |
BRINDLEY PJ, BACHINI M, ILYAS SI, et al. Cholangiocarcinoma[J]. Nat Rev Dis Primers, 2021, 7: 65. DOI: 10.1038/s41572-021-00300-2. |
| [46] |
PINTO C, GIORDANO DM, MARONI L, et al. Role of inflammation and proinflammatory cytokines in cholangiocyte pathophysiology[J]. Biochim Biophys Acta Mol Basis Dis, 2018, 1864( 4 Pt B): 1270- 1278. DOI: 10.1016/j.bbadis.2017.07.024. |
| [47] |
KHAN SA, TAVOLARI S, BRANDI G. Cholangiocarcinoma: Epidemiology and risk factors[J]. Liver Int, 2019, 39( Suppl 1): 19- 31. DOI: 10.1111/liv.14095. |
| [48] |
BARNER-RASMUSSEN N, PUKKALA E, JUSSILA A, et al. Epidemiology, risk of malignancy and patient survival in primary sclerosing cholangitis: A population-based study in Finland[J]. Scand J Gastroenterol, 2020, 55( 1): 74- 81. DOI: 10.1080/00365521.2019.1707277. |
| [49] |
PEREIRA P, AHO V, AROLA J, et al. Bile microbiota in primary sclerosing cholangitis: Impact on disease progression and development of biliary dysplasia[J]. PLoS One, 2017, 12( 8): e0182924. DOI: 10.1371/journal.pone.0182924. |
| [50] |
DYSON JK, BEUERS U, JONES DEJ, et al. Primary sclerosing cholangitis[J]. Lancet, 2018, 391( 10139): 2547- 2559. DOI: 10.1016/s0140-6736(18)30300-3. |
| [51] |
SUTTIPRAPA S, SOTILLO J, SMOUT M, et al. Opisthorchis viverrini proteome and host-parasite interactions[J]. Adv Parasitol, 2018, 102: 45- 72. DOI: 10.1016/bs.apar.2018.06.002. |
| [52] |
XU T, ZHANG B, ZHOU WC. Research progress on the relationship between biliary flora and cholangiocarcinoma[J]. Chin J Bases Clin Gen Surg, 2021, 28( 3): 404- 408. DOI: 10.7507/1007-9424.202006047. |
| [53] |
CHNG KR, CHAN SH, NG AHQ, et al. Tissue microbiome profiling identifies an enrichment of specific enteric bacteria in Opisthorchis viverrini associated cholangiocarcinoma[J]. EBioMedicine, 2016, 8: 195- 202. DOI: 10.1016/j.ebiom.2016.04.034. |
| [54] |
CHEN BR, FU SW, LU LG, et al. A preliminary study of biliary microbiota in patients with bile duct stones or distal cholangiocarcinoma[J]. Biomed Res Int, 2019, 2019: 1092563. DOI: 10.1155/2019/1092563. |







 DownLoad:
DownLoad: