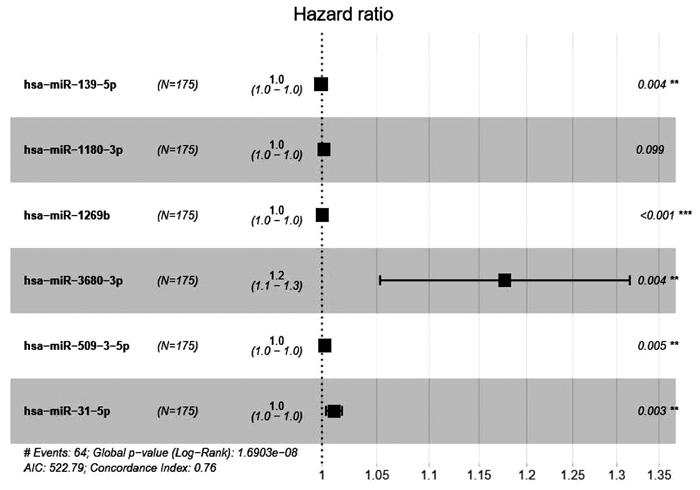
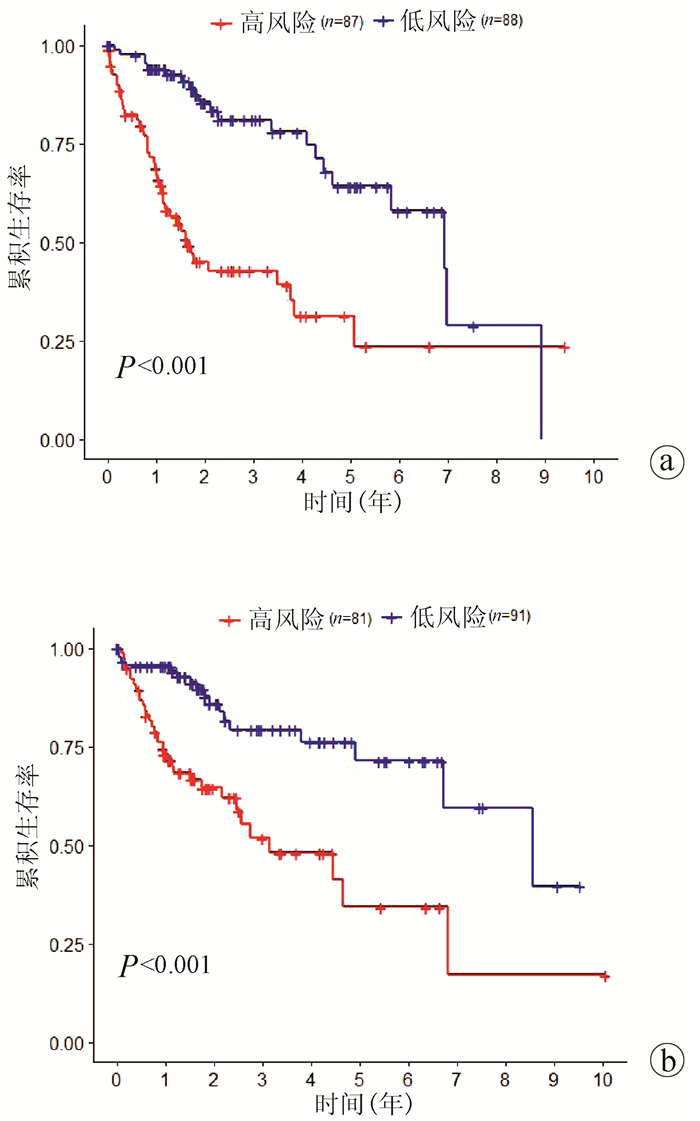
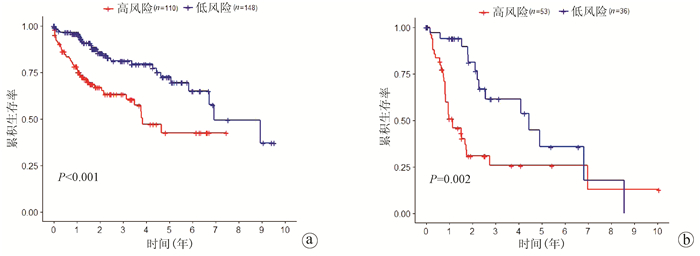
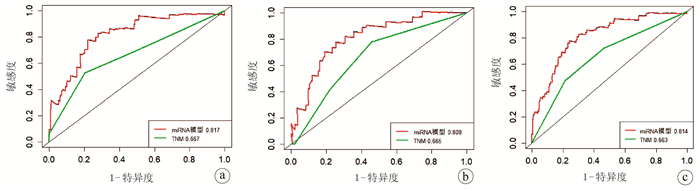
| [1] |
|
| [2] |
VILLANUEVA A. Hepatocellular carcinoma[J]. New Engl J Med, 2019, 380(15): 1450-1462. DOI: 10.1056/NEJMra1713263. |
| [3] |
KULIK L, EL-SERAG HB. Epidemiology and management of hepatocellular carcinoma[J]. Gastroenterology, 2019, 156(2): 477-491. e1. DOI: 10.1053/j.gastro.2018.08.065. |
| [4] |
YANG JD, HAINAUT P, GORES GJ, et al. A global view of hepatocellular carcinoma: Trends, risk, prevention and management[J]. Nat Rev Gastroenterol Hepatol, 2019, 16(10): 589-604. DOI: 10.1038/s41575-019-0186-y. |
| [5] |
LIN S, GREGORY RI. MicroRNA biogenesis pathways in cancer[J]. Nat Rev Cancer, 2015, 15(6): 321-333. DOI: 10.1038/nrc3932. |
| [6] |
RUPAIMOOLE R, SLACK FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases[J]. Nat Rev Drug Discov, 2017, 16(3): 203-222. DOI: 10.1038/nrd.2016.246. |
| [7] |
YERUKALA SATHIPATI S, HO SY. Identifying a miRNA signature for predicting the stage of breast cancer[J]. Sci Rep, 2018, 8(1): 16138. DOI: 10.1038/s41598-018-34604-3. |
| [8] |
KANWAL R, PLAGA AR, LIU X, et al. MicroRNAs in prostate cancer: Functional role as biomarkers[J]. Cancer Lett, 2017, 407: 9-20. DOI: 10.1016/j.canlet.2017.08.011. |
| [9] |
DEB B, UDDIN A, CHAKRABORTY S. miRNAs and ovarian cancer: An overview[J]. J Cell Physiol, 2018, 233(5): 3846-3854. DOI: 10.1002/jcp.26095. |
| [10] |
Bureau of Medical Administration National Health Commission of the People's Republic of China. Guidelines for diagnosis and treatment of primary liver cancer in China (2019 edition)[J]. J Clin Hepatol, 2020, 36(2): 277-292. DOI: 10.3969/j.issn.1001-5256.2020.02.007. |
| [11] |
TOMCZAK K, CZERWIŃSKA P, WIZNEROWICZ M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge[J]. Contemp Oncol (Pozn), 2015, 19(1A): a68-a77. DOI: 10.5114/wo.2014.47136. |
| [12] |
LONG J, ZHANG L, WAN X, et al. A four-gene-based prognostic model predicts overall survival in patients with hepatocellular carcinoma[J]. J Cell Mol Med, 2018, 22(12): 5928-5938. DOI: 10.1111/jcmm.13863. |
| [13] |
WANG X, GAO J, ZHOU B, et al. Identification of prognostic markers for hepatocellular carcinoma based on miRNA expression profiles[J]. Life Sci, 2019, 232: 116596. DOI: 10.1016/j.lfs.2019.116596. |
| [14] |
JIA D, LI S, LI D, et al. Mining TCGA database for genes of prognostic value in glioblastoma microenvironment[J]. Aging (Albany NY), 2018, 10(4): 592-605. DOI: 10.18632/aging.101415. |
| [15] |
LI Y, GU J, XU F, et al. Transcriptomic and functional network features of lung squamous cell carcinoma through integrative analysis of GEO and TCGA data[J]. Sci Rep, 2018, 8(1): 15834. DOI: 10.1038/s41598-018-34160-w. |
| [16] |
NAGY A, LANCZKY A, MENYHART O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets[J]. Sci Rep, 2018, 8(1): 9227. DOI: 10.1038/s41598-018-27521-y. |
| [17] |
TIBSHIRANI R. The lasso method for variable selection in the Cox model[J]. Stat Med, 1997, 16(4): 385-395. DOI: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3.
|
| [18] |
WEI L, WANG X, LV L, et al. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma[J]. Mol Cancer, 2019, 18(1): 147. DOI: 10.1186/s12943-019-1086-z. |
| [19] |
SADRI NAHAND J, BOKHARAEI-SALIM F, SALMANINEJAD A, et al. microRNAs: Key players in virus-associated hepatocellular carcinoma[J]. J Cell Physiol, 2019, 234(8): 12188-12225. DOI: 10.1002/jcp.27956. |
| [20] |
WONG CM, TSANG FH, NG IO. Non-coding RNAs in hepatocellular carcinoma: Molecular functions and pathological implications[J]. Nat Rev Gastroenterol Hepatol, 2018, 15(3): 137-151. DOI: 10.1038/nrgastro.2017.169. |
| [21] |
HUA S, LEI L, DENG L, et al. miR-139-5p inhibits aerobic glycolysis, cell proliferation, migration, and invasion in hepatocellular carcinoma via a reciprocal regulatory interaction with ETS1[J]. Oncogene, 2018, 37(12): 1624-1636. DOI: 10.1038/s41388-017-0057-3. |















 DownLoad:
DownLoad: