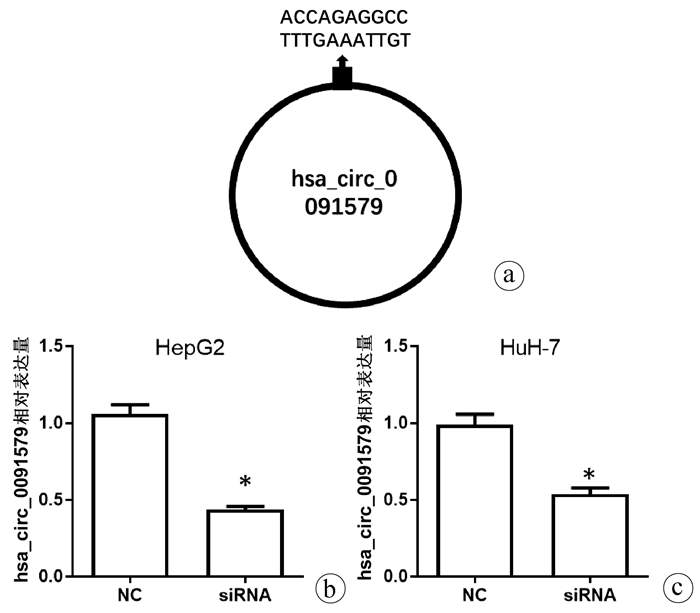
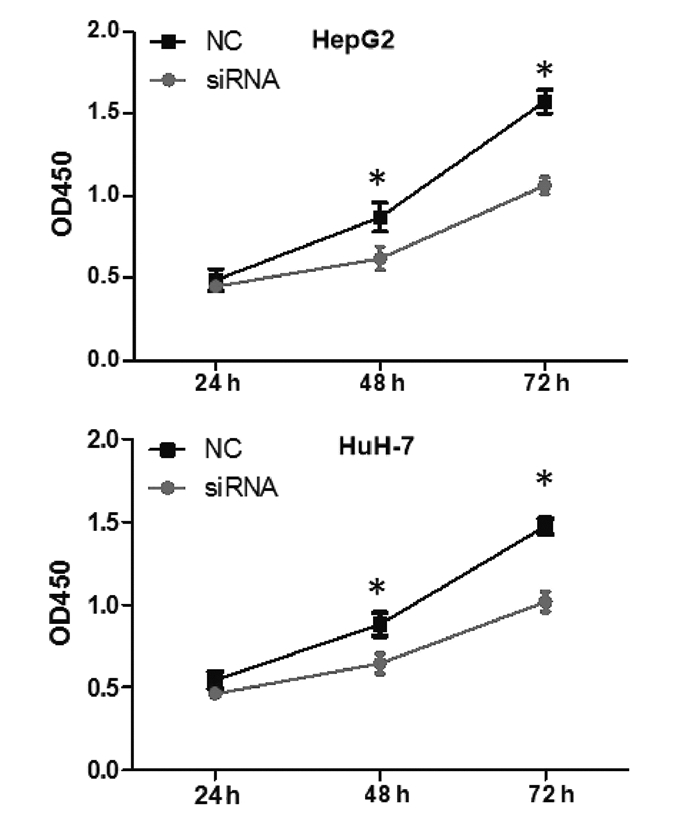
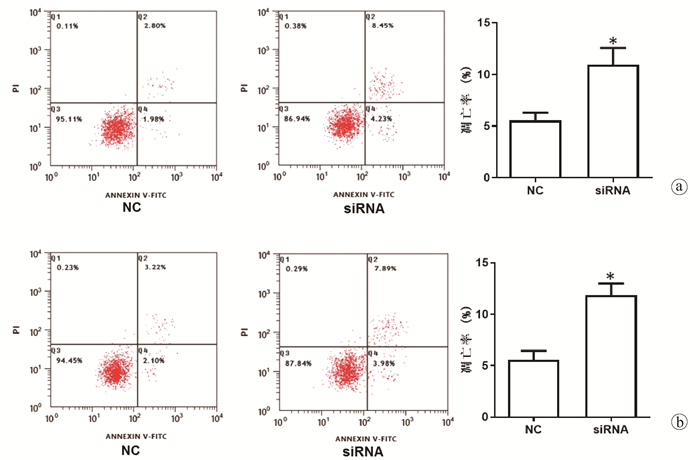
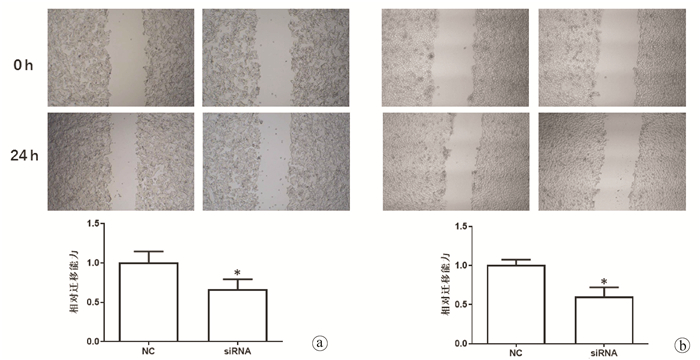
| [1] |
CRAIG AJ, von FELDEN J, GARCIA-LEZANA T, et al. Tumour evolution in hepatocellular carcinoma[J]. Nat Rev Gastroenterol Hepatol, 2020, 17(3): 139-152. DOI: 10.1038/s41575-019-0229-4. |
| [2] |
CHEN F, FANG Y, ZHAO R, et al. Evolution in medicinal chemistry of sorafenib derivatives for hepatocellular carcinoma[J]. Eur J Med Chem, 2019, 179: 916-935. DOI: 10.1016/j.ejmech.2019.06.070. |
| [3] |
QIU M, XIA W, CHEN R, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma[J]. Cancer Res, 2018, 78(11): 2839-2851. DOI: 10.1158/0008-5472.CAN-17-2808. |
| [4] |
TAN S, GOU Q, PU W, et al. Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer[J]. Cell Res, 2018, 28(6): 693-695. DOI: 10.1038/s41422-018-0033-7. |
| [5] |
JECK WR, SHARPLESS NE. Detecting and characterizing circular RNAs[J]. Nat Biotechnol, 2014, 32(5): 453-461. DOI: 10.1038/nbt.2890. |
| [6] |
MEMCZAK S, JENS M, ELEFSINIOTI A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency[J]. Nature, 2013, 495(7441): 333-338. DOI: 10.1038/nature11928. |
| [7] |
TANG Q, HANN SS. Biological roles and mechanisms of circular RNA in human cancers[J]. Onco Targets Ther, 2020, 13: 2067-2092. DOI: 10.2147/OTT.S233672. |
| [8] |
LI J, SUN D, PU W, et al. Circular RNAs in cancer: Biogenesis, function, and clinical significance[J]. Trends Cancer, 2020, 6(4): 319-336. DOI: 10.1016/j.trecan.2020.01.012. |
| [9] |
HUANG A, ZHENG H, WU Z, et al. Circular RNA-protein interactions: Functions, mechanisms, and identification[J]. Theranostics, 2020, 10(8): 3503-3517. DOI: 10.7150/thno.42174. |
| [10] |
HAN B, CHAO J, YAO H. Circular RNA and its mechanisms in disease: From the bench to the clinic[J]. Pharmacol Ther, 2018, 187: 31-44. DOI: 10.1016/j.pharmthera.2018.01.010. |
| [11] |
ZHANG TR, HUANG WQ. Angiogenic circular RNAs: A new landscape in cardiovascular diseases[J]. Microvasc Res, 2020, 129: 103983. DOI: 10.1016/j.mvr.2020.103983. |
| [12] |
XIONG DD, FENG ZB, LAI ZF, et al. High throughput circRNA sequencing analysis reveals novel insights into the mechanism of nitidine chloride against hepatocellular carcinoma[J]. Cell Death Dis, 2019, 10(9): 658. DOI: 10.1038/s41419-019-1890-9. |
| [13] |
ZOU H, XU X, LUO L, et al. Hsa_circ_0101432 promotes the development of hepatocellular carcinoma (HCC) by adsorbing miR-1258 and miR-622[J]. Cell Cycle, 2019, 18(19): 2398-2413. DOI: 10.1080/15384101.2019.1618120. |
| [14] |
WEI X, ZHENG W, TIAN P, et al. Oncogenic hsa_circ_0091581 promotes the malignancy of HCC cell through blocking miR-526b from degrading c-MYC mRNA[J]. Cell Cycle, 2020, 19(7): 817-824. DOI: 10.1080/15384101.2020.1731945. |
| [15] |
ZHANG C, ZHANG C, LIN J, et al. Circular RNA hsa_circ_0091579 serves as a diagnostic and prognostic marker for hepatocellular carcinoma[J]. Cell Physiol Biochem, 2018, 51(1): 290-300. DOI: 10.1159/000495230. |
| [16] |
ZUO Q, HE J, ZHANG S, et al. PPARγ coactivator-1α suppresses metastasis of hepatocellular carcinoma by inhibiting Warburg effect by PPARγ-dependent WNT/β-catenin/pyruvate dehydrogenase kinase isozyme 1 axis[J]. Hepatology, 2021, 73(2): 644-660. DOI: 10.1002/hep.31280. |
| [17] |
CHATURVEDI VK, SINGH A, DUBEY SK, et al. Molecular mechanistic insight of hepatitis B virus mediated hepatocellular carcinoma[J]. Microb Pathog, 2019, 128: 184-194. DOI: 10.1016/j.micpath.2019.01.004. |
| [18] |
XIONG DD, DANG YW, LIN P, et al. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma[J]. J Transl Med, 2018, 16(1): 220. DOI: 10.1186/s12967-018-1593-5. |
| [19] |
O'BRIEN A, ZHOU T, TAN C, et al. Role of non-coding RNAs in the progression of liver cancer: Evidence from experimental models[J]. Cancers (Basel), 2019, 11(11): 1652. DOI: 10.3390/cancers11111652. |
| [20] |
WEI L, WANG X, LV L, et al. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma[J]. Mol Cancer, 2019, 18(1): 147. DOI: 10.1186/s12943-019-1086-z. |
| [21] |
QIU L, XU H, JI M, et al. Circular RNAs in hepatocellular carcinoma: Biomarkers, functions and mechanisms[J]. Life Sci, 2019, 231: 116660. DOI: 10.1016/j.lfs.2019.116660. |
| [22] |
QIU LP, WU YH, YU XF, et al. The emerging role of circular RNAs in hepatocellular carcinoma[J]. J Cancer, 2018, 9(9): 1548-1559. DOI: 10.7150/jca.24566. |
| [23] |
JIANG YL, SHANG MM, DONG SZ, et al. Abnormally expressed circular RNAs as novel non-invasive biomarkers for hepatocellular carcinoma: A meta-analysis[J]. World J Gastrointest Oncol, 2019, 11(10): 909-924. DOI: 10.4251/wjgo.v11.i10.909. |
| [24] |
FU Y, CAI L, LEI X, et al. Circular RNA ABCB10 promotes hepatocellular carcinoma progression by increasing HMG20A expression by sponging miR-670-3p[J]. Cancer Cell Int, 2019, 19: 338. DOI: 10.1186/s12935-019-1055-z. |
| [25] |
CHEN H, LIU S, LI M, et al. circ_0003418 inhibits tumorigenesis and cisplatin chemoresistance through Wnt/β-catenin pathway in hepatocellular carcinoma[J]. Onco Targets Ther, 2019, 12: 9539-9549. DOI: 10.2147/OTT.S229507. |
| [26] |
GURIA A, SHARMA P, NATESAN S, et al. Circular RNAs-the road less traveled[J]. Front Mol Biosci, 2019, 6: 146. DOI: 10.3389/fmolb.2019.00146. |
| [27] |
CHENG JL, LI DJ, LV MY, et al. LncRNA KCNQ1OT1 regulates the invasion and migration of hepatocellular carcinoma by acting on S1PR1 through miR-149[J]. Cancer Gene Ther, 2020. DOI:10.1038/s41417-020-0203-x. [Online ahead of print]
|
| [28] |
CHEN W, YE L, WEN D, et al. MiR-490-5p inhibits hepatocellular carcinoma cell proliferation, migration and invasion by directly regulating ROBO1[J]. Pathol Oncol Res, 2019, 25(1): 1-9. DOI: 10.1007/s12253-017-0305-4. |
| [29] |
CHEN S, LI F, CHAI H, et al. miR-502 inhibits cell proliferation and tumor growth in hepatocellular carcinoma through suppressing phosphoinositide 3-kinase catalytic subunit gamma[J]. Biochem Biophys Res Commun, 2015, 464(2): 500-505. DOI: 10.1016/j.bbrc.2015.06.168. |







 DownLoad:
DownLoad: