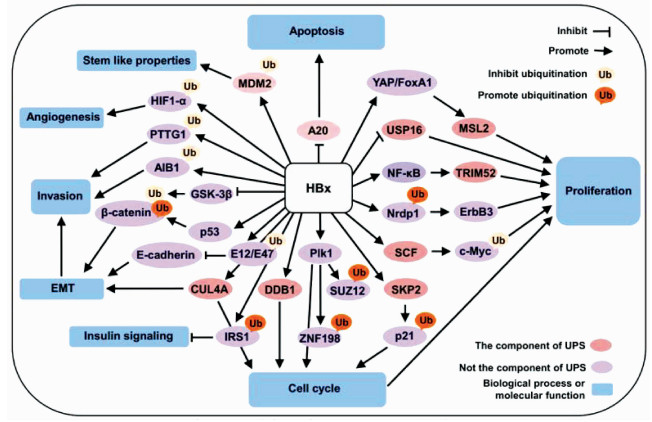
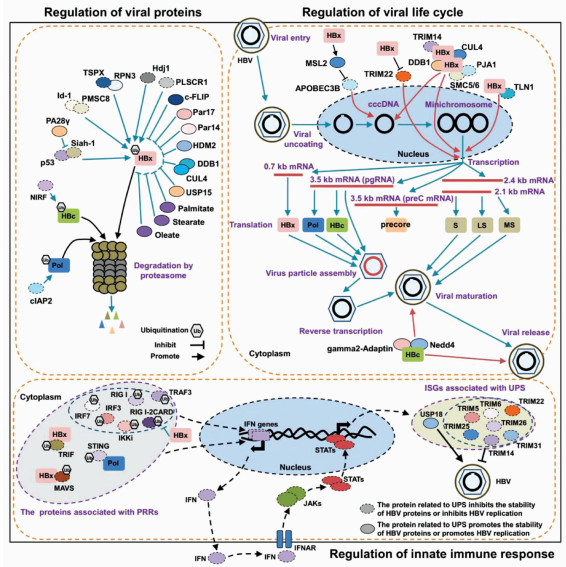
| [1] |
TONG S, REVILL P. Overview of hepatitis B viral replication and genetic variability[J]. J Hepatol, 2016, 64(1 Suppl): s4-s16. DOI: 10.1016/j.jhep.2016.01.027. |
| [2] |
HICKE L. Protein regulation by monoubiquitin[J]. Nat Rev Mol Cell Biol, 2001, 2(3): 195-201. DOI: 10.1038/35056583. |
| [3] |
|
| [4] |
ZHENG Y, GAO C. Fine-tuning of antiviral innate immunity by ubiquitination[J]. Adv Immunol, 2020, 145: 95-128. DOI: 10.1016/bs.ai.2019.11.004. |
| [5] |
TOH QC, TAN TL, TEO WQ, et al. Identification of cellular membrane proteins interacting with hepatitis B surface antigen using yeast split-ubiquitin system[J]. Int J Med Sci, 2005, 2(3): 114-117. DOI: 10.7150/ijms.2.114. |
| [6] |
HARTMANN-STVHLER C, PRANGE R. Hepatitis B virus large envelope protein interacts with gamma2-adaptin, a clathrin adaptor-related protein[J]. J Virol, 2001, 75(11): 5343-5351. DOI: 10.1128/JVI.75.11.5343-5351.2001. |
| [7] |
MANGOLD CM, STREECK RE. Mutational analysis of the cysteine residues in the hepatitis B virus small envelope protein[J]. J Virol, 1993, 67(8): 4588-4597. DOI: 10.1128/JVI.67.8.4588-4597.1993. |
| [8] |
LIU Y, TESTA JS, PHILIP R, et al. A ubiquitin independent degradation pathway utilized by a hepatitis B virus envelope protein to limit antigen presentation[J]. PLoS One, 2011, 6(9): e24477. DOI: 10.1371/journal.pone.0024477. |
| [9] |
SIMSEK E, MEHTA A, ZHOU T, et al. Hepatitis B virus large and middle glycoproteins are degraded by a proteasome pathway in glucosidase-inhibited cells but not in cells with functional glucosidase enzyme[J]. J Virol, 2005, 79(20): 12914-12920. DOI: 10.1128/JVI.79.20.12914-12920.2005. |
| [10] |
ROST M, MANN S, LAMBERT C, et al. Gamma-adaptin, a novel ubiquitin-interacting adaptor, and Nedd4 ubiquitin ligase control hepatitis B virus maturation[J]. J Biol Chem, 2006, 281(39): 29297-29308. DOI: 10.1074/jbc.M603517200. |
| [11] |
GARCIA ML, BYFIELD R, ROBEK MD. Hepatitis B virus replication and release are independent of core lysine ubiquitination[J]. J Virol, 2009, 83(10): 4923-4933. DOI: 10.1128/JVI.02644-08. |
| [12] |
LANGEROVÁ H, LUBYOVÁ B, ZÁBRANSKÝ A, et al. Hepatitis B core protein is post-translationally modified through K29-linked ubiquitination[J]. Cells, 2020, 9(12): 2547. DOI: 10.3390/cells9122547. |
| [13] |
QIAN G, JIN F, CHANG L, et al. NIRF, a novel ubiquitin ligase, interacts with hepatitis B virus core protein and promotes its degradation[J]. Biotechnol Lett, 2012, 34(1): 29-36. DOI: 10.1007/s10529-011-0751-0. |
| [14] |
QIAN G, HU B, ZHOU D, et al. NIRF, a novel ubiquitin ligase, inhibits hepatitis B virus replication through effect on HBV core protein and H3 histones[J]. DNA Cell Biol, 2015, 34(5): 327-332. DOI: 10.1089/dna.2014.2714. |
| [15] |
CHEN JH, YU YS, LIU HH, et al. Ubiquitin conjugation of hepatitis B virus core antigen DNA vaccine leads to enhanced cell-mediated immune response in BALB/c mice[J]. Hepat Mon, 2011, 11(8): 620-628. DOI: 10.5812/kowsar.1735143x.689. |
| [16] |
|
| [17] |
CHEN JH, YU YS, CHEN XH, et al. Enhancement of CTLs induced by DCs loaded with ubiquitinated hepatitis B virus core antigen[J]. World J Gastroenterol, 2012, 18(12): 1319-1327. DOI: 10.3748/wjg.v18.i12.1319. |
| [18] |
SONG L, ZHUO M, TANG Y, et al. Ubiquitin-modified hepatitis B virus core antigen effectively facilitates antigen presentation and enhances cytotoxic T lymphocyte activity via the cytoplasmic transduction peptide in vitro[J]. Mol Med Rep, 2015, 12(1): 289-296. DOI: 10.3892/mmr.2015.3352. |
| [19] |
ZHUO M, SONG L, TANG Y, et al. Vaccination with ubiquitin-hepatitis B core antigen-cytoplasmic transduction peptide enhances the hepatitis B virus-specific cytotoxic T-lymphocyte immune response and inhibits hepatitis B virus replication in transgenic mice[J]. Mol Med Rep, 2015, 12(3): 3591-3598. DOI: 10.3892/mmr.2015.3834. |
| [20] |
SONG L, ZHUO M, TANG Y, et al. Ubiquitin-hepatitis B core antigen-cytoplasmic transduction peptide enhances HBV-specific humoral and CTL immune responses in vivo[J]. Int Immunopharmacol, 2014, 23(1): 1-7. DOI: 10.1016/j.intimp.2014.08.006. |
| [21] |
WANG Y, CUI L, YANG G, et al. Hepatitis B e antigen inhibits NF-κB activity by interrupting K63-linked ubiquitination of NEMO[J]. J Virol, 2019, 93(2): e00667-18. DOI: 10.1128/JVI.00667-18. |
| [22] |
HOU L, ZHAO J, GAO S, et al. Restriction of hepatitis B virus replication by c-Abl-induced proteasomal degradation of the viral polymerase[J]. Sci Adv, 2019, 5(2): eaau7130. DOI: 10.1126/sciadv.aau7130. |
| [23] |
LIU X, SHAO J, XIONG W, et al. Cellular cIAP2 gene expression associated with anti-HBV activity of TNF-alpha in hepatoblastoma cells[J]. J Interferon Cytokine Res, 2005, 25(10): 617-626. DOI: 10.1089/jir.2005.25.617. |
| [24] |
WANG Z, NI J, LI J, et al. Inhibition of hepatitis B virus replication by cIAP2 involves accelerating the ubiquitin-proteasome-mediated destruction of polymerase[J]. J Virol, 2011, 85(21): 11457-11467. DOI: 10.1128/JVI.00879-11. |
| [25] |
MU T, ZHAO X, ZHU Y, et al. The E3 ubiquitin ligase TRIM21 promotes HBV DNA polymerase degradation[J]. Viruses, 2020, 12(3): 346. DOI: 10.3390/v12030346. |
| [26] |
LIU Y, LI J, CHEN J, et al. Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways[J]. J Virol, 2015, 89(4): 2287-2300. DOI: 10.1128/JVI.02760-14. |
| [27] |
HU Z, ZHANG Z, DOO E, et al. Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex[J]. J Virol, 1999, 73(9): 7231-7240. DOI: 10.1128/JVI.73.9.7231-7240.1999. |
| [28] |
CHEN H, ZHANG Y, YE S, et al. Chromatin remodelling factor BAF155 protects hepatitis B virus X protein (HBx) from ubiquitin-independent proteasomal degradation[J]. Emerg Microbes Infect, 2019, 8(1): 1393-1405. DOI: 10.1080/22221751.2019.1666661. |
| [29] |
ZHANG JF, XIONG HL, CAO JL, et al. A cell-penetrating whole molecule antibody targeting intracellular HBx suppresses hepatitis B virus via TRIM21-dependent pathway[J]. Theranostics, 2018, 8(2): 549-562. DOI: 10.7150/thno.20047. |
| [30] |
ZHAO J, WU J, CAI H, et al. E3 ubiquitin ligase Siah-1 is down-regulated and fails to target natural HBx truncates for degradation in hepatocellular carcinoma[J]. J Cancer, 2016, 7(4): 418-426. DOI: 10.7150/jca.13019. |
| [31] |
YEOM S, KIM SS, JEONG H, et al. Hepatitis B virus X protein activates E3 ubiquitin ligase Siah-1 to control virus propagation via a negative feedback loop[J]. J Gen Virol, 2017, 98(7): 1774-1784. DOI: 10.1099/jgv.0.000856. |
| [32] |
YOO YS, PARK YJ, LEE HS, et al. Mitochondria ubiquitin ligase, MARCH5 resolves hepatitis B virus X protein aggregates in the liver pathogenesis[J]. Cell Death Dis, 2019, 10(12): 938. DOI: 10.1038/s41419-019-2175-z. |
| [33] |
ZHANG H, HUANG C, WANG Y, et al. Hepatitis B virus X protein sensitizes TRAIL-induced hepatocyte apoptosis by inhibiting the E3 ubiquitin ligase A20[J]. PLoS One, 2015, 10(5): e0127329. DOI: 10.1371/journal.pone.0127329. |
| [34] |
MOLINA-JIMÉNEZ F, BENEDICTO I, MURATA M, et al. Expression of pituitary tumor-transforming gene 1 (PTTG1)/securin in hepatitis B virus (HBV)-associated liver diseases: Evidence for an HBV X protein-mediated inhibition of PTTG1 ubiquitination and degradation[J]. Hepatology, 2010, 51(3): 777-787. DOI: 10.1002/hep.23468. |
| [35] |
LEE S, KIM W, KO C, et al. Hepatitis B virus X protein enhances Myc stability by inhibiting SCF(Skp2) ubiquitin E3 ligase-mediated Myc ubiquitination and contributes to oncogenesis[J]. Oncogene, 2016, 35(14): 1857-1867. DOI: 10.1038/onc.2015.251. |
| [36] |
GUO L, WANG X, REN L, et al. HBx affects CUL4-DDB1 function in both positive and negative manners[J]. Biochem Biophys Res Commun, 2014, 450(4): 1492-1497. DOI: 10.1016/j.bbrc.2014.07.019. |
| [37] |
JIANG J, TANG H. Mechanism of inhibiting type I interferon induction by hepatitis B virus X protein[J]. Protein Cell, 2010, 1(12): 1106-1117. DOI: 10.1007/s13238-010-0141-8. |
| [38] |
WEI C, NI C, SONG T, et al. The hepatitis B virus X protein disrupts innate immunity by downregulating mitochondrial antiviral signaling protein[J]. J Immunol, 2010, 185(2): 1158-1168. DOI: 10.4049/jimmunol.0903874. |
| [39] |
KONG F, YOU H, KONG D, et al. The interaction of hepatitis B virus with the ubiquitin proteasome system in viral replication and associated pathogenesis[J]. Virol J, 2019, 16(1): 73. DOI: 10.1186/s12985-019-1183-z. |
| [40] |
CHEN J, WU M, LIU K, et al. New insights into hepatitis B virus biology and implications for novel antiviral strategies[J]. National Science Review, 2015, 2(4): 296-313. DOI: 10.1093/nsr/nwv044. |
| [41] |
ZHOU L, REN JH, CHENG ST, et al. A functional variant in ubiquitin conjugating enzyme E2 L3 contributes to hepatitis B virus infection and maintains covalently closed circular DNA stability by inducing degradation of apolipoprotein B mRNA editing enzyme catalytic subunit 3A[J]. Hepatology, 2019, 69(5): 1885-1902. DOI: 10.1002/hep.30497. |
| [42] |
WANG Z, KAWAGUCHI K, HONDA M, et al. Notch signaling facilitates hepatitis B virus covalently closed circular DNA transcription via cAMP response element-binding protein with E3 ubiquitin ligase-modulation[J]. Sci Rep, 2019, 9(1): 1621. DOI: 10.1038/s41598-018-38139-5. |
| [43] |
CHENG ST, HU JL, REN JH, et al. Dicoumarol, an NQO1 inhibitor, blocks cccDNA transcription by promoting degradation of HBx[J]. J Hepatol, 2021, 74(3): 522-534. DOI: 10.1016/j.jhep.2020.09.019. |
| [44] |
DECORSIÈRE A, MUELLER H, van BREUGEL PC, et al. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor[J]. Nature, 2016, 531(7594): 386-389. DOI: 10.1038/nature17170. |
| [45] |
MURPHY CM, XU Y, LI F, et al. Hepatitis B virus X protein promotes degradation of SMC5/6 to enhance HBV replication[J]. Cell Rep, 2016, 16(11): 2846-2854. DOI: 10.1016/j.celrep.2016.08.026. |



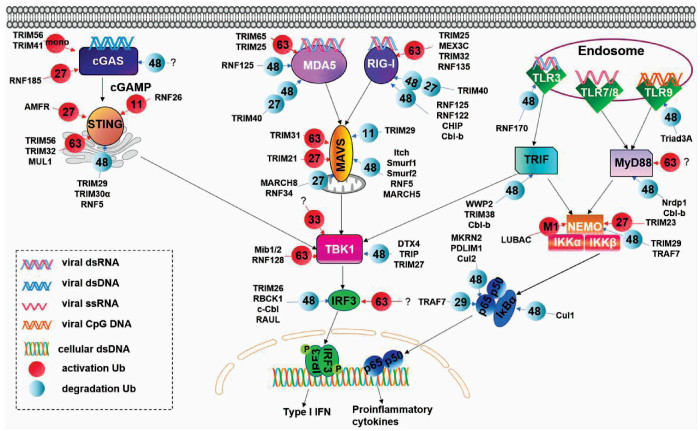




 DownLoad:
DownLoad: