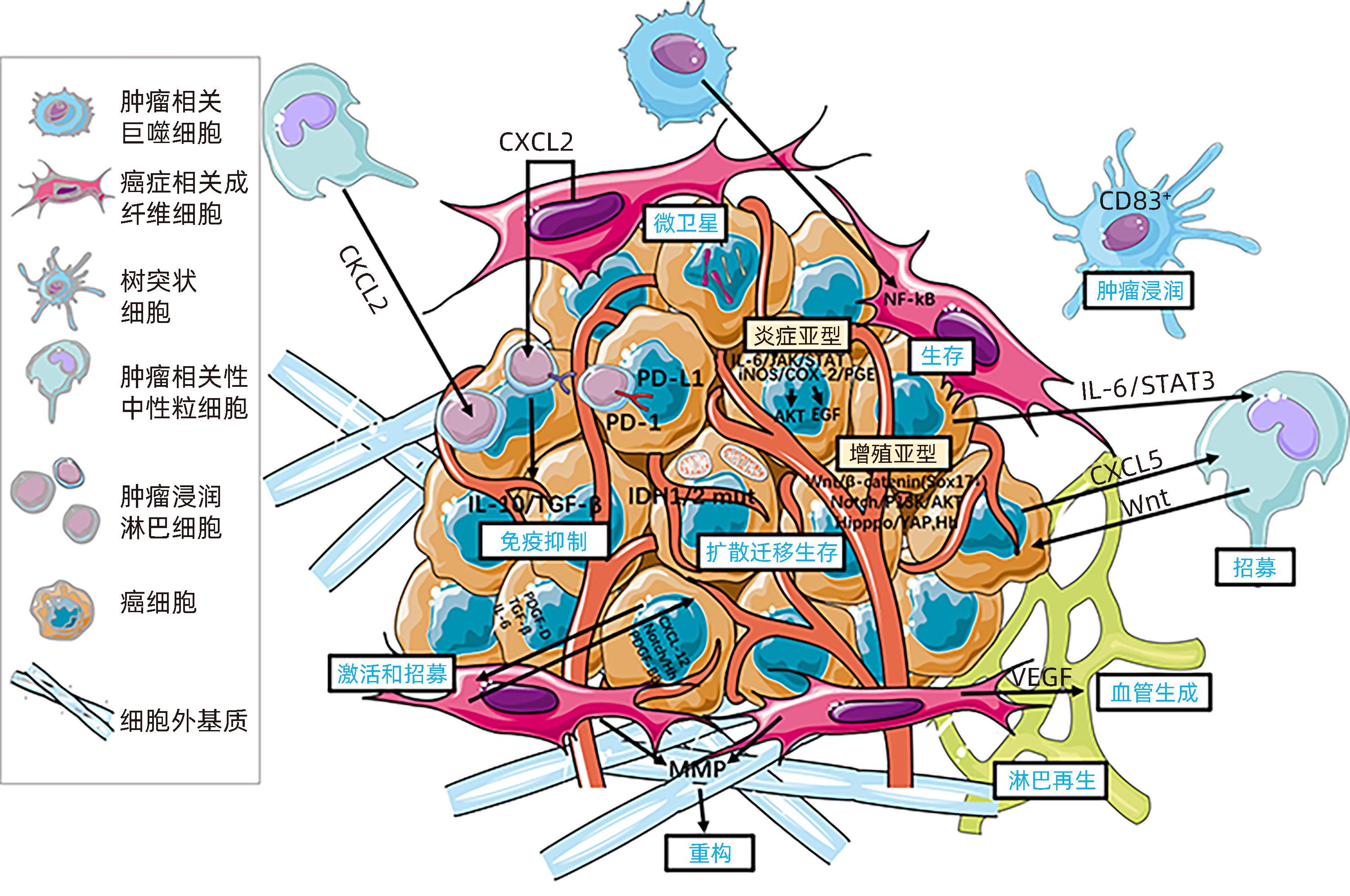
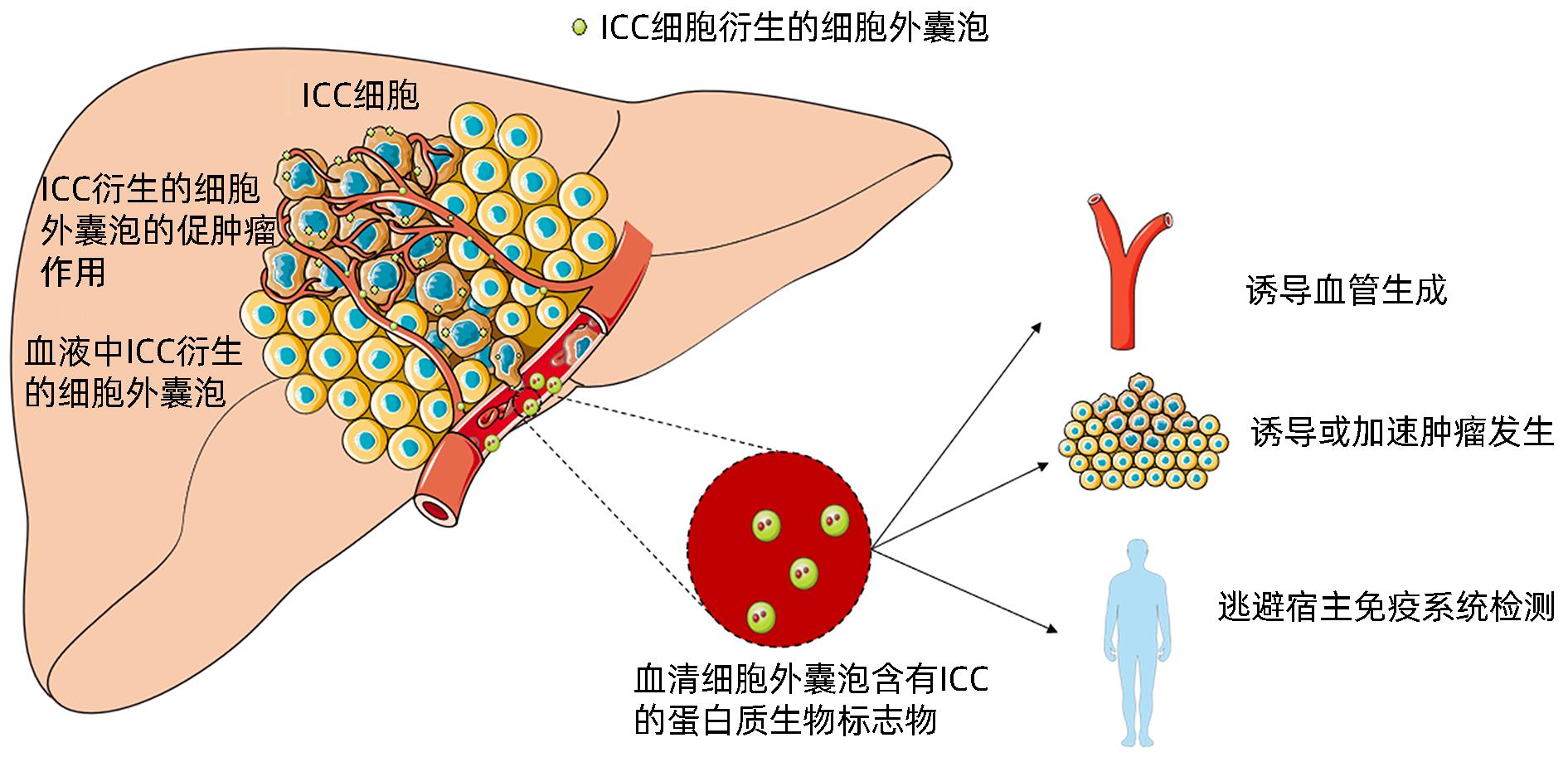
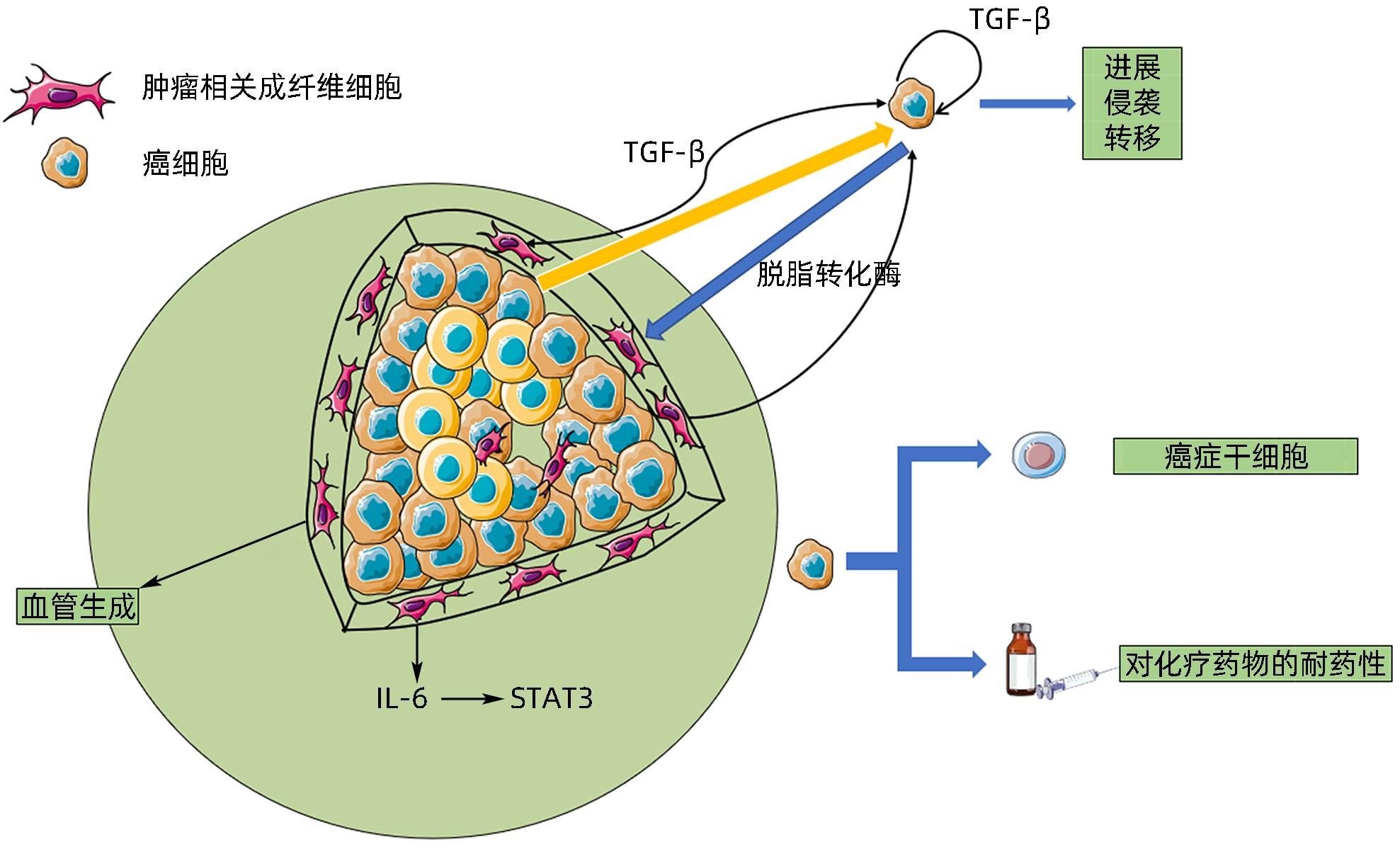
| [1] |
YANG KG, WANG WW, WANG Y, et al. Proteomic analysis of serum and serum exosomes, and their application in intrahepatic cholangiocarcinoma[J]. Chin J Chromatogr, 2021, 39( 11): 1191- 1202. DOI: 10.3724/SP.J.1123.2021.04009. |
| [2] |
CAO LP, HONG JW, WU J. Potential of extracellular vesicles and exosomes as diagnostic markers for cholangiocarcinoma[J]. Hepatobiliary Surg Nutr, 2022, 11( 3): 436- 438. DOI: 10.21037/hbsn-2022-02. |
| [3] |
ARAI Y, TOTOKI Y, HOSODA F, et al. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma[J]. Hepatology, 2014, 59( 4): 1427- 1434. DOI: 10.1002/hep.26890. |
| [4] |
SIA D, HOSHIDA Y, VILLANUEVA A, et al. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes[J]. Gastroenterology, 2013, 144( 4): 829- 840. DOI: 10.1053/j.gastro.2013.01.001. |
| [5] |
ANDERSEN JB, SPEE B, BLECHACZ BR, et al. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors[J]. Gastroenterology, 2012, 142( 4): 1021- 1031.e15. DOI: 10.1053/j.gastro.2011.12.005. |
| [6] |
NEPAL C, O’ROURKE CJ, OLIVEIRA DVNP, et al. Genomic perturbations reveal distinct regulatory networks in intrahepatic cholangiocarcinoma[J]. Hepatology, 2018, 68( 3): 949- 963. DOI: 10.1002/hep.29764. |
| [7] |
LIN JZ, SHI JP, GUO HL, et al. Alterations in DNA damage repair genes in primary liver cancer[J]. Clin Cancer Res, 2019, 25( 15): 4701- 4711. DOI: 10.1158/1078-0432.CCR-19-0127. |
| [8] |
MAYNARD H, STADLER ZK, BERGER MF, et al. Germline alterations in patients with biliary tract cancers: A spectrum of significant and previously underappreciated findings[J]. Cancer, 2020, 126( 9): 1995- 2002. DOI: 10.1002/cncr.32740. |
| [9] |
JUSAKUL A, CUTCUTACHE I, YONG CH, et al. Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma[J]. Cancer Discov, 2017, 7( 10): 1116- 1135. DOI: 10.1158/2159-8290.CD-17-0368. |
| [10] |
CHAN-ON W, NAIRISMÄGI ML, ONG CK, et al. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers[J]. Nat Genet, 2013, 45( 12): 1474- 1478. DOI: 10.1038/ng.2806. |
| [11] |
ONG CK, SUBIMERB C, PAIROJKUL C, et al. Exome sequencing of liver fluke-associated cholangiocarcinoma[J]. Nat Genet, 2012, 44( 6): 690- 693. DOI: 10.1038/ng.2273. |
| [12] |
|
| [13] |
XING C, LI H, LI RJ, et al. The roles of exosomal immune checkpoint proteins in tumors[J]. Mil Med Res, 2021, 8( 1): 56. DOI: 10.1186/s40779-021-00350-3. |
| [14] |
HELWA I, CAI JW, DREWRY MD, et al. A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents[J]. PLoS One, 2017, 12( 1): e0170628. DOI: 10.1371/journal.pone.0170628. |
| [15] |
LOBB RJ, BECKER M, WEN SW, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma[J]. J Extracell Vesicles, 2015, 4: 27031. DOI: 10.3402/jev.v4.27031. |
| [16] |
HMMIER A, O’BRIEN ME, LYNCH V, et al. Proteomic analysis of bronchoalveolar lavage fluid(BALF) from lung cancer patients using label-free mass spectrometry[J]. BBA Clin, 2017, 7: 97- 104. DOI: 10.1016/j.bbacli.2017.03.001. |
| [17] |
YÁÑEZ-MÓ M, SILJANDER PRM, ANDREU Z, et al. Biological properties of extracellular vesicles and their physiological functions[J]. J Extracell Vesicles, 2015, 4: 27066. DOI: 10.3402/jev.v4.27066. |
| [18] |
GUO J, ZHONG XX, TAN QL, et al. miR-301a-3p induced by endoplasmic reticulum stress mediates the occurrence and transmission of trastuzumab resistance in HER2-positive gastric cancer[J]. Cell Death Dis, 2021, 12( 7): 696. DOI: 10.1038/s41419-021-03991-3. |
| [19] |
ZHOU DJ, GE W, CAO DD. Advances in the study of exosomes in the development and diagnosis of hepatocellular liver cancer[J]. Chin Med Herald, 2023, 20( 10): 42- 44, 54. DOI: 10.20047/j.issn1673-7210.2023.10.08. |
| [20] |
WEERAPHAN C, PHONGDARA A, CHAIYAWAT P, et al. Phosphoproteome profiling of isogenic cancer cell-derived exosome reveals HSP90 as a potential marker for human cholangiocarcinoma[J]. Proteomics, 2019, 19( 12): e1800159. DOI: 10.1002/pmic.201800159. |
| [21] |
IKEDA C, HAGA H, MAKINO N, et al. Utility of Claudin-3 in extracellular vesicles from human bile as biomarkers of cholangiocarcinoma[J]. Sci Rep, 2021, 11( 1): 1195. DOI: 10.1038/s41598-021-81023-y. |
| [22] |
ARBELAIZ A, AZKARGORTA M, KRAWCZYK M, et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma[J]. Hepatology, 2017, 66( 4): 1125- 1143. DOI: 10.1002/hep.29291. |
| [23] |
PAN Y, SHAO SJ, SUN H, et al. Bile-derived exosome noncoding RNAs as potential diagnostic and prognostic biomarkers for cholangiocarcinoma[J]. Front Oncol, 2022, 12: 985089. DOI: 10.3389/fonc.2022.985089. |
| [24] |
HAN JY, AHN KS, KIM YH, et al. Circulating microRNAs as biomarkers in bile-derived exosomes of cholangiocarcinoma[J]. Ann Surg Treat Res, 2021, 101( 3): 140- 150. DOI: 10.4174/astr.2021.101.3.140. |
| [25] |
GAO LL, YANG XP, ZHANG H, et al. Inhibition of miR-10a-5p suppresses cholangiocarcinoma cell growth through downregulation of Akt pathway[J]. Onco Targets Ther, 2018, 11: 6981- 6994. DOI: 10.2147/OTT.S182225. |
| [26] |
LI O, JIANG B, YI WM, et al. LncRNA NEAT1 promotes cell proliferation, migration, and invasion via the miR-186-5p/PTP4A1 axis in cholangiocarcinoma[J]. Kaohsiung J Med Sci, 2021, 37( 5): 379- 391. DOI: 10.1002/kjm2.12354. |
| [27] |
SUN ZP, TAN ZG, PENG C, et al. LncRNA SNHG3 facilitates the malignant phenotype of cholangiocarcinoma cells via the miR-3173-5p/ERG axis[J]. J Gastrointest Surg, 2022, 26( 4): 802- 812. DOI: 10.1007/s11605-021-05160-5. |
| [28] |
CHEN Q, WANG HB, LI Z, et al. Circular RNA ACTN 4 promotes intrahepatic cholangiocarcinoma progression by recruiting YBX1 to initiate FZD7 transcription[J]. J Hepatol, 2022, 76( 1): 135- 147. DOI: 10.1016/j.jhep.2021.08.027. |
| [29] |
CHEN HW, CHENGALVALA V, HU HX, et al. Tumor-derived exosomes: Nanovesicles made by cancer cells to promote cancer metastasis[J]. Acta Pharm Sin B, 2021, 11( 8): 2136- 2149. DOI: 10.1016/j.apsb.2021.04.012. |
| [30] |
LIU JY, REN LW, LI S, et al. The biology, function, and applications of exosomes in cancer[J]. Acta Pharm Sin B, 2021, 11( 9): 2783- 2797. DOI: 10.1016/j.apsb.2021.01.001. |
| [31] |
LI L, ZHAO J, ZHANG QB, et al. Cancer cell-derived exosomes promote HCC tumorigenesis through hedgehog pathway[J]. Front Oncol, 2021, 11: 756205. DOI: 10.3389/fonc.2021.756205. |
| [32] |
WANG YZ, YI J, CHEN XG, et al. The regulation of cancer cell migration by lung cancer cell-derived exosomes through TGF-β and IL-10[J]. Oncol Lett, 2016, 11( 2): 1527- 1530. DOI: 10.3892/ol.2015.4044. |
| [33] |
LI XX, WANG SH, ZHU RJ, et al. Lung tumor exosomes induce a pro-inflammatory phenotype in mesenchymal stem cells via NFκB-TLR signaling pathway[J]. J Hematol Oncol, 2016, 9: 42. DOI: 10.1186/s13045-016-0269-y. |
| [34] |
LIN LY, DU LM, CAO K, et al. Tumour cell-derived exosomes endow mesenchymal stromal cells with tumour-promotion capabilities[J]. Oncogene, 2016, 35( 46): 6038- 6042. DOI: 10.1038/onc.2016.131. |
| [35] |
BECKER A, THAKUR BK, WEISS JM, et al. Extracellular vesicles in cancer: Cell-to-cell mediators of metastasis[J]. Cancer Cell, 2016, 30( 6): 836- 848. DOI: 10.1016/j.ccell.2016.10.009. |
| [36] |
HE LQ, ZHU W, CHEN Q, et al. Ovarian cancer cell-secreted exosomal miR-205 promotes metastasis by inducing angiogenesis[J]. Theranostics, 2019, 9( 26): 8206- 8220. DOI: 10.7150/thno.37455. |
| [37] |
TANG MKS, YUE PYK, IP PP, et al. Soluble E-cadherin promotes tumor angiogenesis and localizes to exosome surface[J]. Nat Commun, 2018, 9( 1): 2270. DOI: 10.1038/s41467-018-04695-7. |
| [38] |
PARK JE, DUTTA B, TSE SW, et al. Hypoxia-induced tumor exosomes promote M2-like macrophage polarization of infiltrating myeloid cells and microRNA-mediated metabolic shift[J]. Oncogene, 2019, 38( 26): 5158- 5173. DOI: 10.1038/s41388-019-0782-x. |
| [39] |
ZHANG PF, GAO C, HUANG XY, et al. Cancer cell-derived exosomal circUHRF 1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma[J]. Mol Cancer, 2020, 19( 1): 110. DOI: 10.1186/s12943-020-01222-5. |
| [40] |
XIE FT, XU MX, LU J, et al. The role of exosomal PD-L1 in tumor progression and immunotherapy[J]. Mol Cancer, 2019, 18( 1): 146. DOI: 10.1186/s12943-019-1074-3. |
| [41] |
YIN Z, YU M, MA TT, et al. Mechanisms underlying low-clinical responses to PD-1/PD-L1 blocking antibodies in immunotherapy of cancer: A key role of exosomal PD-L1[J]. J Immunother Cancer, 2021, 9( 1): e001698. DOI: 10.1136/jitc-2020-001698. |
| [42] |
MOON B, CHANG S. Exosome as a delivery vehicle for cancer therapy[J]. Cells, 2022, 11( 3): 316. DOI: 10.3390/cells11030316. |
| [43] |
WALKER S, BUSATTO S, PHAM A, et al. Extracellular vesicle-based drug delivery systems for cancer treatment[J]. Theranostics, 2019, 9( 26): 8001- 8017. DOI: 10.7150/thno.37097. |
| [44] |
LI L, PIONTEK K, ISHIDA M, et al. Extracellular vesicles carry microRNA-195 to intrahepatic cholangiocarcinoma and improve survival in a rat model[J]. Hepatology, 2017, 65( 2): 501- 514. DOI: 10.1002/hep.28735. |
| [45] |
SHA M, JEONG S, QIU BJ, et al. Isolation of cancer-associated fibroblasts and its promotion to the progression of intrahepatic cholangiocarcinoma[J]. Cancer Med, 2018, 7( 9): 4665- 4677. DOI: 10.1002/cam4.1704. |
| [46] |
SIRICA AE, CAMPBELL DJ, DUMUR CI. Cancer-associated fibroblasts in intrahepatic cholangiocarcinoma[J]. Curr Opin Gastroenterol, 2011, 27( 3): 276- 284. DOI: 10.1097/MOG.0b013e32834405c3. |
| [47] |
FABRIS L, SATO K, ALPINI G, et al. The tumor microenvironment in cholangiocarcinoma progression[J]. Hepatology, 2021, 73( Suppl 1): 75- 85. DOI: 10.1002/hep.31410. |
| [48] |
YANG RJ, WANG D, HAN S, et al. Erratum: miR-206 suppresses the deterioration of intrahepatic cholangiocarcinoma and promotes sensitivity to chemotherapy by inhibiting interactions with stromal CAF: Erratum[J]. Int J Biol Sci, 2022, 18( 11): 4466- 4467. DOI: 10.7150/ijbs.75760. |
| [49] |
OTA Y, TAKAHASHI K, OTAKE S, et al. Extracellular vesicle-encapsulated miR-30e suppresses cholangiocarcinoma cell invasion and migration via inhibiting epithelial-mesenchymal transition[J]. Oncotarget, 2018, 9( 23): 16400- 16417. DOI: 10.18632/oncotarget.24711. |



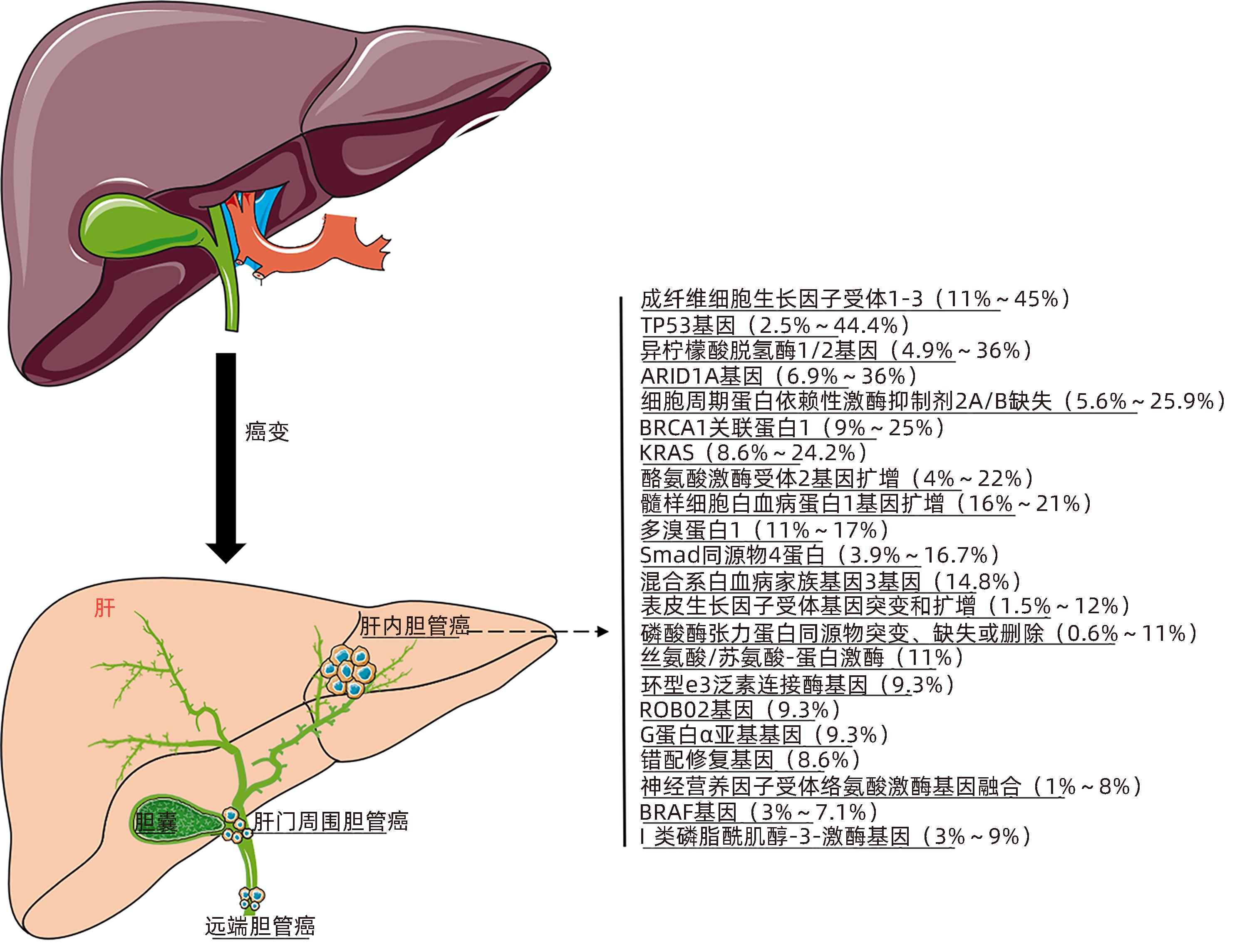




 DownLoad:
DownLoad: