| [1] |
World Health Organization. Global hepatitis report[R]. Geneva: WHO, 2017: 1-83.
|
| [2] |
|
| [3] |
SUMMERS J, MASON WS. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate[J]. Cell, 1982, 29(2): 403-415. DOI: 10.1016/0092-8674(82)90157-x. |
| [4] |
HU J, PROTZER U, SIDDIQUI A. Revisiting hepatitis B virus: Challenges of curative therapies[J]. J Virol, 2019, 93(20): e01032-19. DOI: 10.1128/JVI.01032-19. |
| [5] |
GUO JT, GUO H. Metabolism and function of hepatitis B virus cccDNA: Implications for the development of cccDNA-targeting antiviral therapeutics[J]. Antiviral Res, 2015, 122: 91-100. DOI: 10.1016/j.antiviral.2015.08.005. |
| [6] |
ZHU A, LIAO X, LI S, et al. HBV cccDNA and its potential as a therapeutic target[J]. J Clin Transl Hepatol, 2019, 7(3): 258-262. DOI: 10.14218/JCTH.2018.00054. |
| [7] |
LIN CL, KAO JH. Review article: Novel therapies for hepatitis B virus cure - advances and perspectives[J]. Aliment Pharmacol Ther, 2016, 44(3): 213-222. DOI: 10.1111/apt.13694. |
| [8] |
LIU S, XIN Y. HBV cccDNA: The stumbling block for treatment of HBV infection[J]. J Clin Transl Hepatol, 2019, 7(3): 195-196. DOI: 10.14218/JCTH.2019.00047. |
| [9] |
TANG L, COVERT E, WILSON E, et al. Chronic hepatitis B infection: A review[J]. JAMA, 2018, 319(17): 1802-1813. DOI: 10.1001/jama.2018.3795. |
| [10] |
BARTENSCHLAGER R, URBAN S, PROTZER U. Towards curative therapy of chronic viral hepatitis[J]. Z Gastroenterol, 2019, 57(1): 61-73. DOI: 10.1055/a-0824-1576. |
| [11] |
BLOCK TM, GUO H, GUO JT. Molecular virology of hepatitis B virus for clinicians[J]. Clin Liver Dis, 2007, 11(4): 685-706, vii. DOI: 10.1016/j.cld.2007.08.002. |
| [12] |
DONG J, YING J, QIU X, et al. Advanced strategies for eliminating the cccDNA of HBV[J]. Dig Dis Sci, 2018, 63(1): 7-15. DOI: 10.1007/s10620-017-4842-1. |
| [13] |
SETO WK, LO YR, PAWLOTSKY JM, et al. Chronic hepatitis B virus infection[J]. Lancet, 2018, 392(10161): 2313-2324. DOI: 10.1016/S0140-6736(18)31865-8. |
| [14] |
TU T, BUDZINSKA MA, VONDRAN F, et al. Hepatitis B virus DNA integration occurs early in the viral life cycle in an in vitro infection model via sodium taurocholate cotransporting polypeptide-dependent uptake of enveloped virus particles[J]. J Virol, 2018, 92(11): e02007-e02017. DOI: 10.1128/JVI.02007-17. |
| [15] |
HU J, SEEGER C. Hepadnavirus genome replication and persistence[J]. Cold Spring Harb Perspect Med, 2015, 5(7): a021386. DOI: 10.1101/cshperspect.a021386. |
| [16] |
SEEGER C, MASON WS. Molecular biology of hepatitis B virus infection[J]. Virology, 2015, 479-480: 672-686. DOI: 10.1016/j.virol.2015.02.031. |
| [17] |
NASSAL M. HBV cccDNA: Viral persistence reservoir and key obstacle for a cure of chronic hepatitis B[J]. Gut, 2015, 64(12): 1972-1984. DOI: 10.1136/gutjnl-2015-309809. |
| [18] |
GAO W, HU J. Formation of hepatitis B virus covalently closed circular DNA: Removal of genome-linked protein[J]. J Virol, 2007, 81(12): 6164-6174. DOI: 10.1128/JVI.02721-06. |
| [19] |
WU M, LI J, YUE L, et al. Establishment of Cre-mediated HBV recombinant cccDNA (rcccDNA) cell line for cccDNA biology and antiviral screening assays[J]. Antiviral Res, 2018, 152: 45-52. DOI: 10.1016/j.antiviral.2018.02.007. |
| [20] |
BLOOM K, MAEPA MB, ELY A, et al. Gene therapy for chronic HBV-can we eliminate cccDNA?[J]. Genes (Basel), 2018, 9(4). DOI: 10.3390/genes9040207. |
| [21] |
KITAMURA K, QUE L, SHIMADU M, et al. Flap endonuclease 1 is involved in cccDNA formation in the hepatitis B virus[J]. PLoS Pathog, 2018, 14(6): e1007124. DOI: 10.1371/journal.ppat.1007124. |
| [22] |
SLAGLE BL, BOUCHARD MJ. Role of HBx in hepatitis B virus persistence and its therapeutic implications[J]. Curr Opin Virol, 2018, 30: 32-38. DOI: 10.1016/j.coviro.2018.01.007. |
| [23] |
SEKIBA K, OTSUKA M, OHNO M, et al. Inhibition of HBV transcription from cccDNA with nitazoxanide by targeting the HBx-DDB1 interaction[J]. Cell Mol Gastroenterol Hepatol, 2019, 7(2): 297-312. DOI: 10.1016/j.jcmgh.2018.10.010. |
| [24] |
LEWANDOWSKA M, PIEKARSKA A. New directions in hepatitis B therapy research[J]. Clin Exp Hepatol, 2017, 3(3): 119-126. DOI: 10.5114/ceh.2017.68831. |
| [25] |
KENNEDY EM, KORNEPATI AV, CULLEN BR. Targeting hepatitis B virus cccDNA using CRISPR/Cas9[J]. Antiviral Res, 2015, 123: 188-192. DOI: 10.1016/j.antiviral.2015.10.004. |
| [26] |
SEEGER C, SOHN JA. Complete spectrum of CRISPR/Cas9-induced mutations on HBV cccDNA[J]. Mol Ther, 2016, 24(7): 1258-1266. DOI: 10.1038/mt.2016.94. |
| [27] |
YANG HC, CHEN PJ. The potential and challenges of CRISPR-Cas in eradication of hepatitis B virus covalently closed circular DNA[J]. Virus Res, 2018, 244: 304-310. DOI: 10.1016/j.virusres.2017.06.010. |
| [28] |
KOSTYUSHEV D, BREZGIN S, KOSTYUSHEVA A, et al. Orthologous CRISPR/Cas9 systems for specific and efficient degradation of covalently closed circular DNA of hepatitis B virus[J]. Cell Mol Life Sci, 2019, 76(9): 1779-1794. DOI: 10.1007/s00018-019-03021-8. |
| [29] |
WEBER ND, STONE D, SEDLAK RH, et al. AAV-mediated delivery of zinc finger nucleases targeting hepatitis B virus inhibits active replication[J]. PLoS One, 2014, 9(5): e97579. DOI: 10.1371/journal.pone.0097579. |
| [30] |
BLOOM K, ELY A, MUSSOLINO C, et al. Inactivation of hepatitis B virus replication in cultured cells and in vivo with engineered transcription activator-like effector nucleases[J]. Mol Ther, 2013, 21(10): 1889-1897. DOI: 10.1038/mt.2013.170. |
| [31] |
DREYER T, NICHOLSON S, ELY A, et al. Improved antiviral efficacy using TALEN-mediated homology directed recombination to introduce artificial primary miRNAs into DNA of hepatitis B virus[J]. Biochem Biophys Res Commun, 2016, 478(4): 1563-1568. DOI: 10.1016/j.bbrc.2016.08.152. |
| [32] |
BELLONI L, ALLWEISS L, GUERRIERI F, et al. IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome[J]. J Clin Invest, 2012, 122(2): 529-537. DOI: 10.1172/JCI58847. |
| [33] |
PARK HK, MIN BY, KIM NY, et al. Short hairpin RNA induces methylation of hepatitis B virus covalently closed circular DNA in human hepatoma cells[J]. Biochem Biophys Res Commun, 2013, 436(2): 152-155. DOI: 10.1016/j.bbrc.2013.04.108. |
| [34] |
QIAN G, HU B, ZHOU D, et al. NIRF, a novel ubiquitin ligase, inhibits hepatitis B virus replication through effect on HBV core protein and H3 histones[J]. DNA Cell Biol, 2015, 34(5): 327-332. DOI: 10.1089/dna.2014.2714. |
| [35] |
WEI ZQ, ZHANG YH, KE CZ, et al. Curcumin inhibits hepatitis B virus infection by down-regulating cccDNA-bound histone acetylation[J]. World J Gastroenterol, 2017, 23(34): 6252-6260. DOI: 10.3748/wjg.v23.i34.6252. |
| [36] |
SONG M, SUN Y, TIAN J, et al. Silencing retinoid X receptor alpha expression enhances early-stage hepatitis B virus infection in cell cultures[J]. J Virol, 2018, 92(8). DOI: 10.1128/JVI.01771-17. |
| [37] |
YUAN Y, ZHAO K, YAO Y, et al. HDAC11 restricts HBV replication through epigenetic repression of cccDNA transcription[J]. Antiviral Res, 2019, 172: 104619. DOI: 10.1016/j.antiviral.2019.104619. |
| [38] |
FANNING GC, ZOULIM F, HOU J, et al. Therapeutic strategies for hepatitis B virus infection: Towards a cure[J]. Nat Rev Drug Discov, 2019, 18(11): 827-844. DOI: 10.1038/s41573-019-0037-0. |
| [39] |
ZAI WJ, CHEN JL, YUAN ZH. Regulatory mechanisms of the transcription and metabolism of hepatitis B virus covalently closed circular DNA and strategies for silencing and elimination[J]. J Clin Hepatol, 2020, 36(5): 983-988. DOI: 10.3969/j.issn.1001-5256.2020.05.006. |









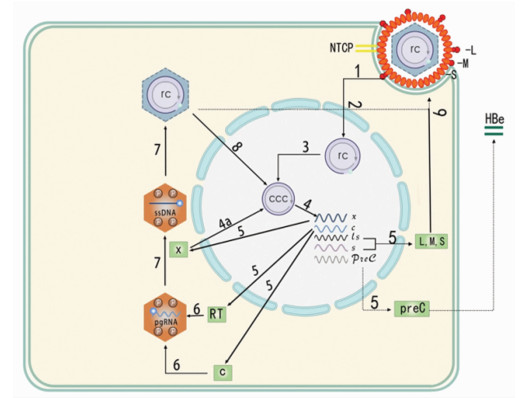




 DownLoad:
DownLoad: